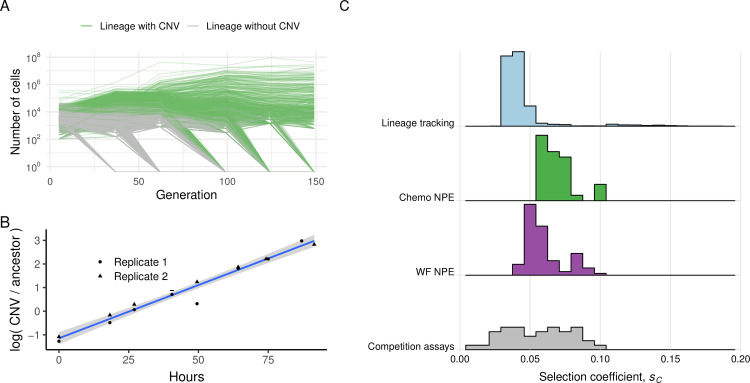
Fig 7. Comparison of DFE inferred using NPE, lineage-tracking barcodes, and competition assays.
(A) Barcode-based lineage frequency trajectories in experimental population bc01. Lineages with (green) and without (gray) GAP1 CNVs are shown. (B) Two replicates of a pairwise competition assay for a single GAP1 CNV containing lineage isolated from an evolving population. The selection coefficient for the clone is estimated from the slope of the linear model (blue line) and 95% CI (gray). (C) The DFE for all beneficial GAP1 CNVs inferred from 11 populations using NPE and the WF (purple) and chemostat (Chemo; green) models compared with the DFE inferred from barcode frequency trajectories in the bc01 population (light blue) and the DFE inferred using pairwise competition assays with different GAP1 CNV containing clones (gray). Data and code required to generate this figure can be found at https://doi.org/10.17605/OSF.IO/E9D5X. CNV, copy number variant; DFE, distribution of fitness effects; NPE, Neural Posterior Estimation; WF, Wright–Fisher.