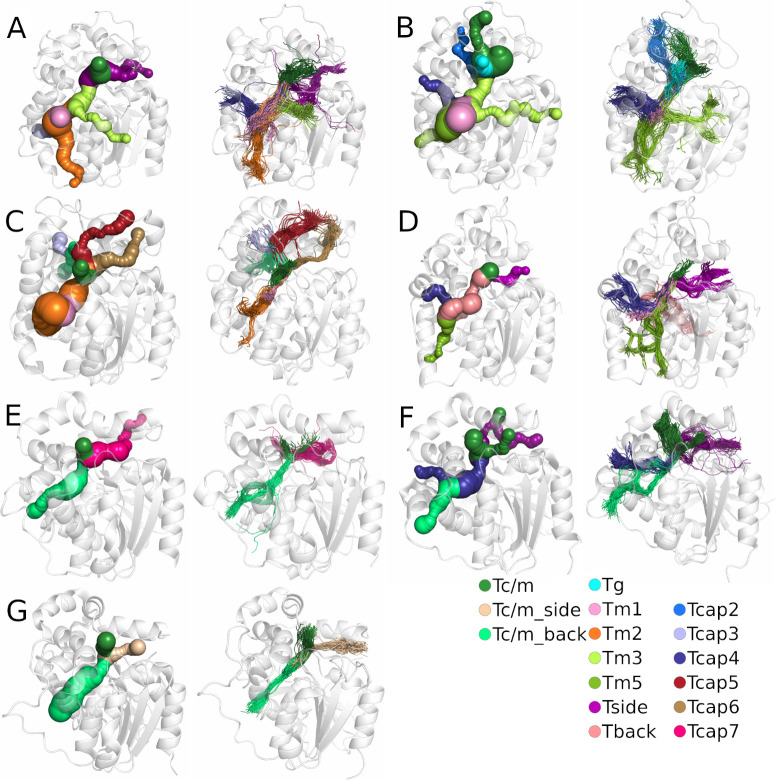
Fig 2.
Comparison of tunnels identified in the crystal structure (left) and the results after molecular dynamics (MD) simulation (right) for each system: A) M. musculus soluble epoxide hydrolase (msEH), B) H. sapiens soluble epoxide hydrolase (hsEH), C) S. tuberosum soluble epoxide hydrolase (StEH1), D) T. reesei soluble epoxide hydrolase (TrEH), E) B. megaterium soluble epoxide hydrolase (bmEH), and thermophilic soluble epoxide hydrolases from an unknown source organism F) Sibe-EH, and G) CH65-EH. Protein structures are shown as white transparent cartoons. Matching tunnels are marked with the same colour as spheres (in crystal structures) and lines (in MD simulations).