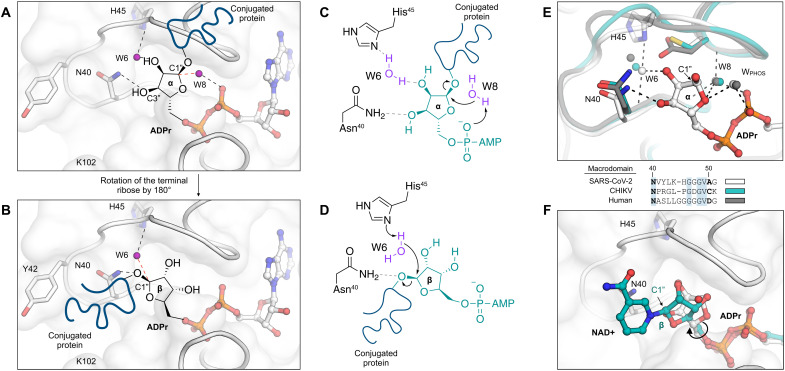
Fig. 8. Mechanism of ADPr-ribose hydrolysis catalyzed by the SARS-CoV-2 NSP3 macrodomain.
(A) Composite image showing the ADPr-bound Mac1 structure (white sticks/cartoon/surface, PDB 7KQP). ADPr is shown in the conformation that is compatible with a substrate-assisted mechanism. The terminal ribose adopts the α epimer, and W8 acts as the water nucleophile. For clarity, only the side chains of selected residues are shown. Hydrogen bonds are shown with dashed black lines, and the W6-C1″ trajectory is shown with a dashed red line. (B) Same as (A) but showing the β epimer of the terminal ribose that is compatible with His45 acting as a general base to activate W6 as a nucleophile. (C and D) Chemical structures showing the two possible mechanisms for ADPr hydrolysis. (E) Top: Structural alignment of the SARS-CoV-2 macrodomain (PDB code 7KQP), Chikungunya virus macrodomain (PDB code 3GPO), and the human macrodomain hMacroD2 (PDB code 4IQY), all in complex with ADPr. Bottom: Protein sequence alignment of residues equivalent to residues 40 to 51 from the SARS-CoV-2 macrodomain. (F) X-ray crystal structure of the NSP3 macrodomain from the Tylonycteris bat coronavirus HKU4 in complex with NAD+ (PDB code 6MEB). The terminal ribose is rotated ~180° relative to ADPr. This configuration matches the model proposed in (B) and (D).