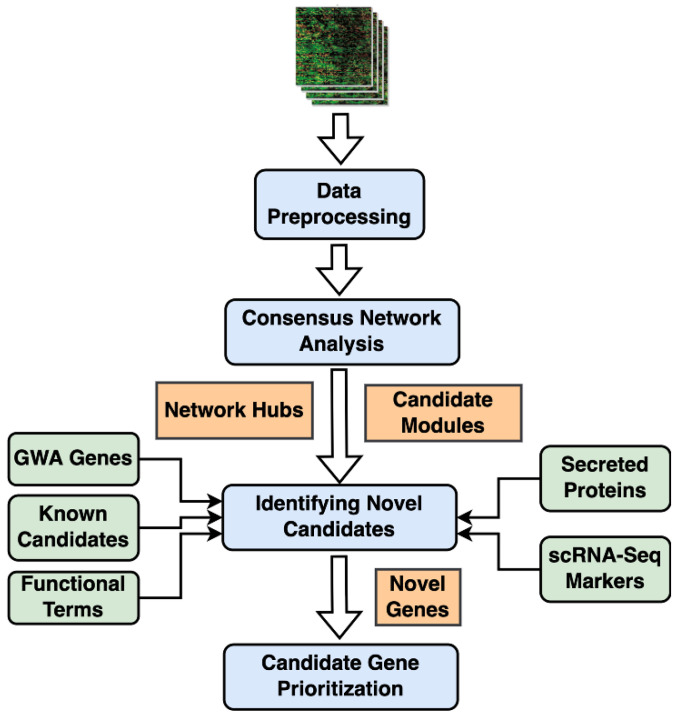
Figure 7.
Schematic representation of the workflow: Transcriptomic data from the IPF and controls were downloaded from NCBI’s Gene Expression Omnibus (GEO) and used as inputs for WGCNA consensus Analysis after the necessary data cleaning and normalization steps. Consensus modules obtained from WGCNA were correlated with phenotype status and other IPF-relevant clinical traits to select candidate modules. Intramodular hub genes were identified within each candidate module and used for functional characterization and to identify novel candidates and secreted biomarkers in IPF.