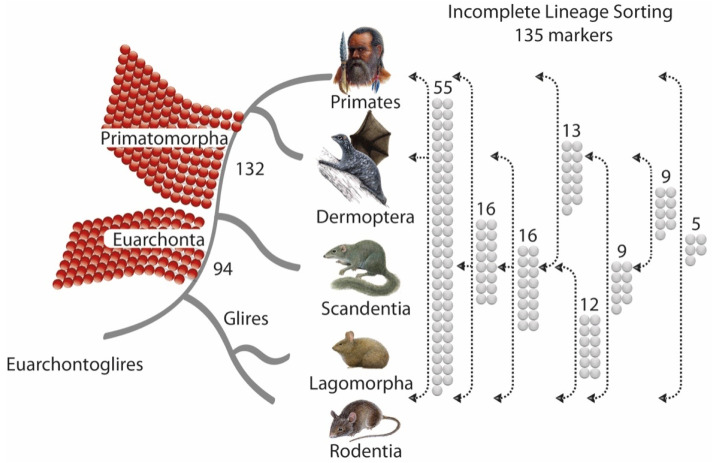
Figure 3.
Phylogenetic reconstruction derived from TE insertion presence/absence patterns and statistically analyzed using the 4-LIN tool. Primatomorpha received the most presence/absence support (132 TE insertions), followed by Euarchonta by 94 diagnostic TEs. All markers supporting conflicting tree topologies (together 135) are indicated as grey balls with respective numbers and relationships. Branches assigned by black arrowheads show the presence of shared grey balls (orthologous insertions). All other species represent their absence. For example, nine TE insertions were present in Dermoptera and Scandentia and absent in Primates and Glires. Lagomorpha only accounted for 299 (including ten rodent-lagomorph conflicting patterns) of the 361 TE markers shown here (for details see text). The complete presence/absence matrix is given as Supplementary Table S1. We did not screen for monophyly markers of Glires and Euarchontoglires, monophyly markers of euarchontogliran orders, or phylogenetic signals within them.