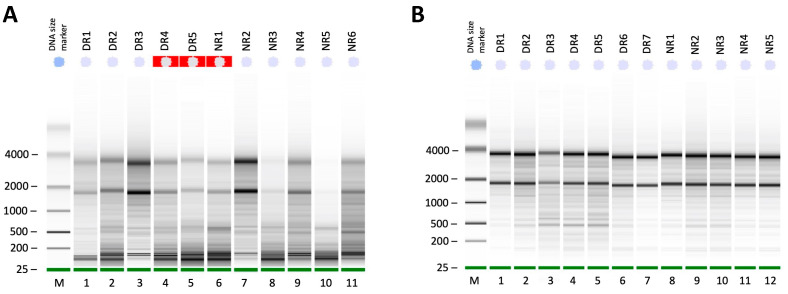
Figure 5.
Virtual gel-like images generated by an Agilent 2100 Bioanalyzer using the pico and nano kits. (A) There was a high level of RNA degradation in samples extracted using the standard RNeasy mini protocol (Qiagen). Samples isolated from diabetic rats (DR) are presented in lanes 1–3, which had an RIN range from 4.2–7, while those from normal rats (NR) in lanes 7–11 had an RIN value between 2.3–7.3. Red highlighted lanes 4–6 show samples with low RIN values that could not be calculated due to degradation or sample contamination. (B) Intact RNA with very low levels of degradation in samples extracted using our optimized method. Samples isolated from diabetic rats (DR) are presented in lanes 1–7, which had an RIN value range from 6.7–8.8, while those from normal rats (NR) in lanes 8–12 had an RIN value ≥8.0 except for NR2 and NR3 (RIN = 7.4, 7.2), which was due to a technical difficulty during the homogenization step.