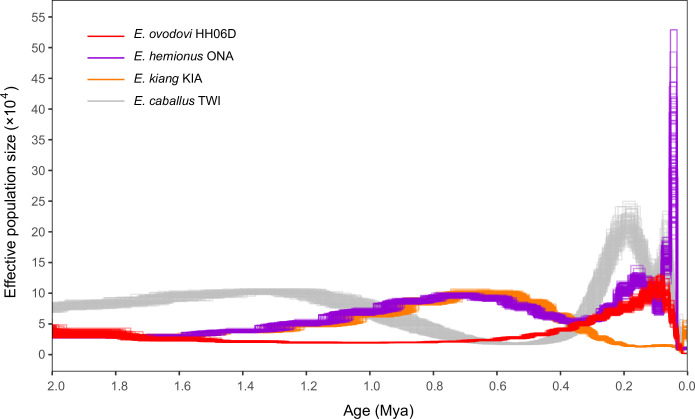
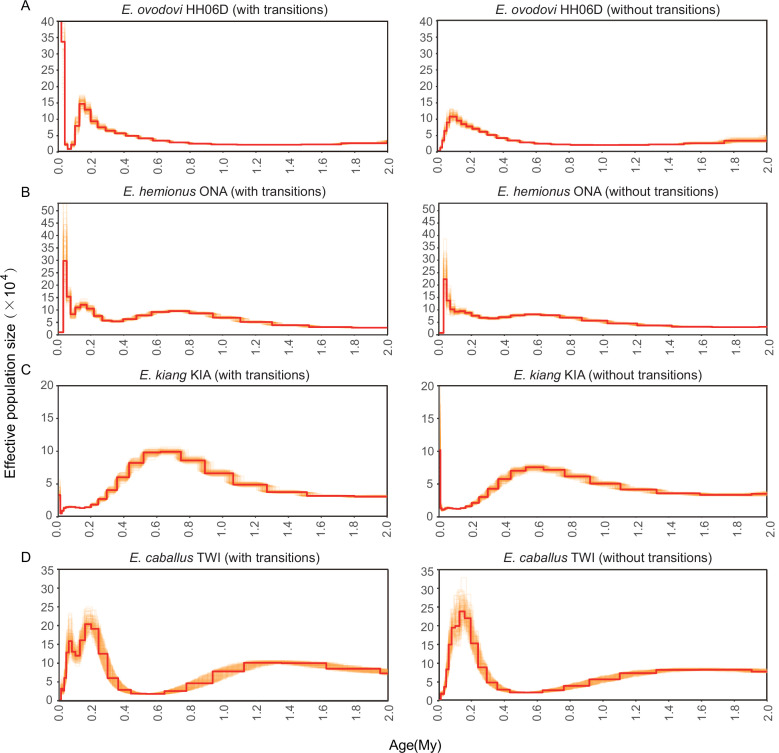
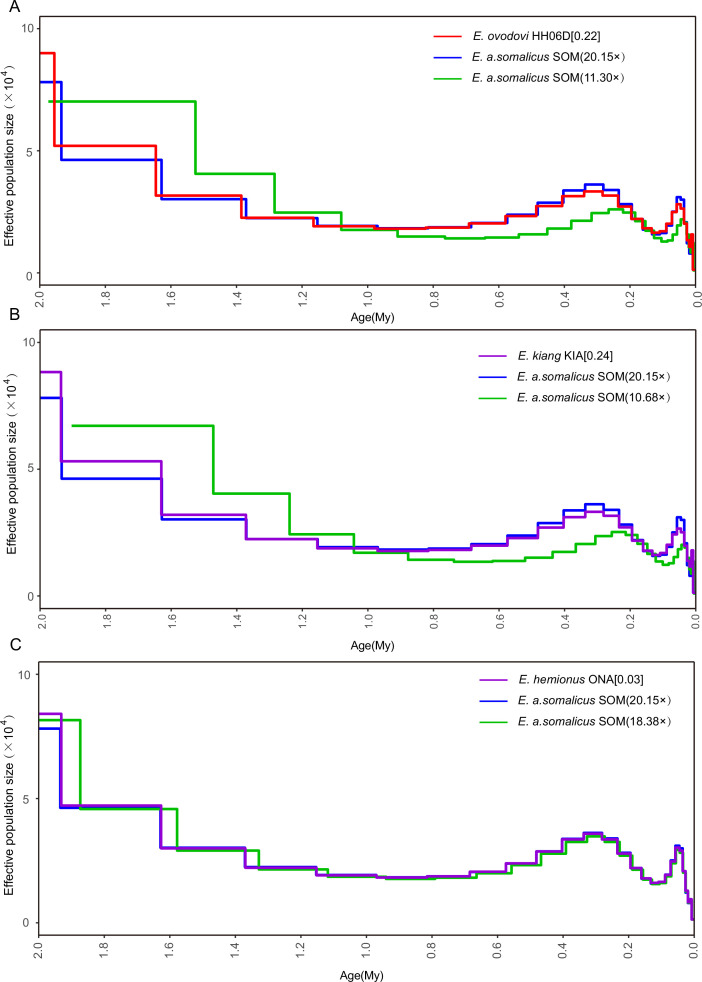
Figure 4. Pairwise sequential Markovian coalescent (PSMC) profiles (100 bootstrap pseudo-replicates) of four Eurasian equine species (E. ovodovi HH06D, E. caballus TWI [Kalbfleisch et al., 2018], E. hemionus ONA, and E. kiang KIA) (Jónsson et al., 2014).
The y-axis represents the effective population size (×10,000), and the x-axis is scaled in millions of years before present. Faded lines show bootstrap values.