Figure 2.
Agent-based model of glioblastoma patient tumor tissue slices infected with oHSV-1
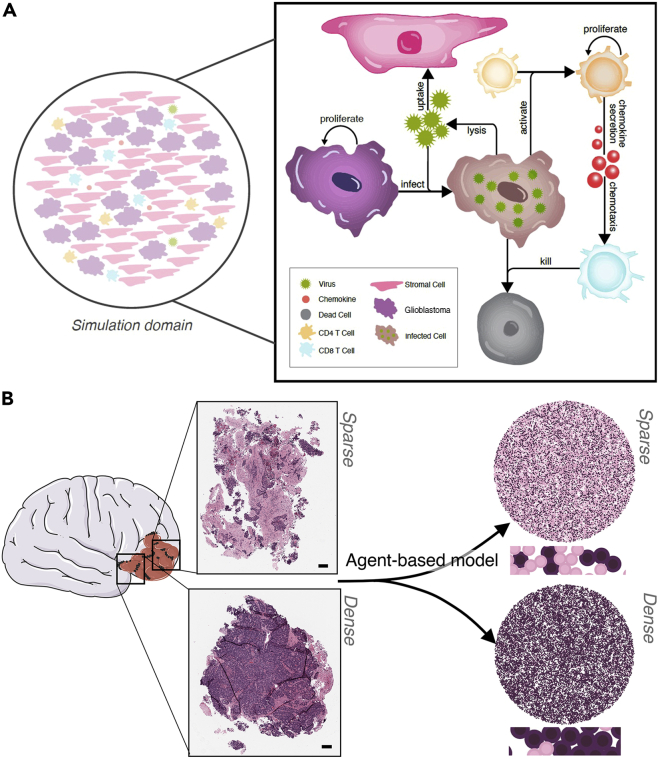
(A) Schematic describing the model agents: glioblastoma (GBM) cell, stromal cell, CD4+ T cell, and CD8+ T cell. Corresponding agent rules and cell–cell interactions are in Table 1. Glioblastoma (purple) and stromal (pink) cells were randomly distributed in a 2D domain inside a circle of radius with initial numbers depending on the stromal cell to glioblastoma cell ratio. oHSV-1 particles were modeled to diffuse through the domain (Equation 1, STAR Methods), bind, and become internalized by glioblastoma or stromal cells. In infected glioblastoma cells (brown cells), the virus then undergoes replication and eventually lyses the infected cell releasing new viral particles and killing the cell (black cell) (Figure S4). CD4+ T cells and CD8+ T cells are both present in the tissue and contribute to localized clearance of infected cells. Stromal cells act as virus sinks. Through contact with an infected tumor cell, a naïve CD4+ T cells (Th) will become activated and secrete chemokines which attract CD8+ T cells (CTL) through chemotaxis. CD8+T cells kill infected cells they encounter (Figure S5).
(B) A patient-derived resected glioblastoma was sub-sectioned into two smaller slices, which were then formalin-fixed, paraffin-embedded, sectioned onto slides, and stained with hematoxylin and eosin (H&E). A clinical pathologist scored the fraction of tumor cell, necrosis, immune cell, and stroma content (Table 2). Scale bar further zoomed in inserts can be found in Figure S6. The two subsections were recreated in our agent-based model and designated as sparse (top) or dense (bottom) based on the pathology scores. Larger versions of the simulations can be found in Figure S7.