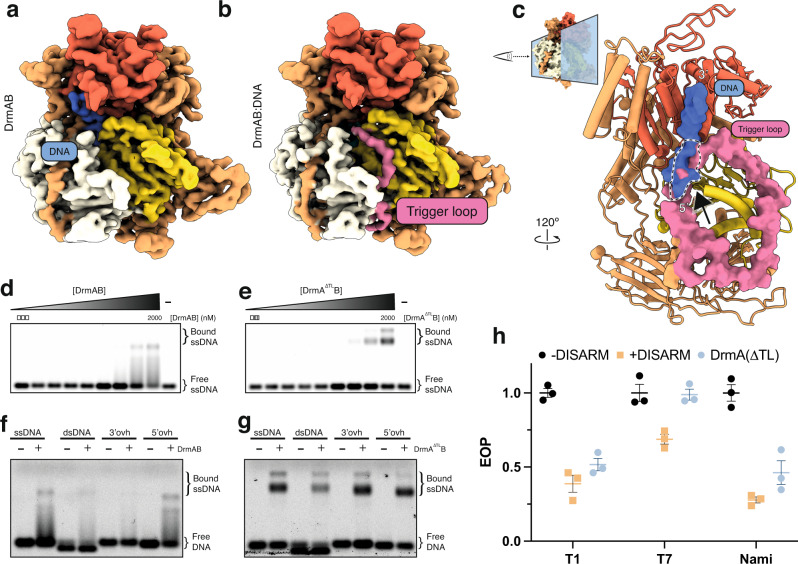
Fig. 3. DrmA trigger loop (TL) partially occludes DNA-binding site.
a, b Cryo-EM reconstruction of apo DrmAB (a) and DNA-bound DrmAB (b). Density corresponding to DrmA trigger loop is shown in pink. DrmAB are colored by structural domains as shown in Fig. 1a. c Overlay of DrmAB-bound DNA (blue) and TL (pink) showing the steric clash (dashed black & white outline). TL partially occludes DNA-binding site. DrmB and parts of DrmA have been omitted for clarity. Graphic at the top right shows how the view in (c) is related to the structures in panels (a, b). d, e EMSA analysis of DrmAB and DrmA(∆loop)B binding DNA. The smeared bands that occur at high concentrations of DrmAB correspond to bound DNA dissociating from the complex. This did not occur for DrmA(∆loop)B, and an additional super-shift was present, corresponding to multiple copies of DrmA(∆loop)B binding to the same 75-nt DNA concurrently. The free DNA marker is 75-nt in length. Representative of three independent experiments. Source Data are provided as a Source Data file. f, g DNA substrate preferences of DrmAB and DrmA(∆loop)B. The 75-nt Cy5-labeled DNA (ssDNA) was annealed to complementary oligos corresponding to the full sequence (dsDNA), or 20 bases at the 5’ or 3’ ends (3’ovh and 5’ovh, respectively). DrmAB showed a preference for ssDNA and 5’ovh DNA, whereas DrmA(∆TL)B bound to all DNA substrates tested. The free DNA marker is 75-nt in length. Representative of three independent experiments. Source Data are provided as a Source Data file. h Effect of truncation of TL on DISARM anti-phage activity. Points correspond to three biological replicates, with mean and standard deviation shown. Source Data are provided as a Source Data file.