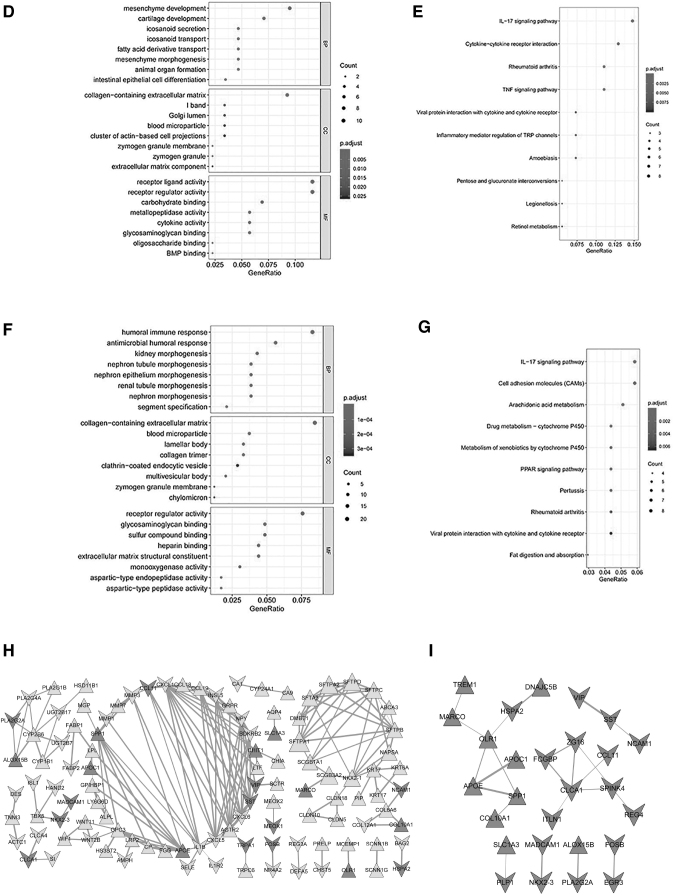
FIG. 4.
Construction of the TF-miRNA-mRNA and PPI network network of immune-related DEGs of M0 macrophage cells. To analyze the TF-miRNA-mRNA network, the miRNAs of the immune-related DEGs were predicted. To analyze the interactions between proteins and proteins encoded by DEGs, the STRING database was utilized to analyze their interactions. (A) The TF-miRNA-mRNA network; (B) the top 8 terms of the GO analysis of upregulated DEGs; (C) the top 8 terms of the KEGG pathway analysis of upregulated DEGs; (D) the top 8 terms of the GO analysis of downregulated DEGs; (E) the top 10 terms of the KEGG pathway analysis of downregulated DEGs; (F) the top 8 terms of the GO analysis of all DEGs; (G) the top 10 terms of the KEGG pathway analysis of all DEGs; (H) the PPI network of the upregulated and downregulated DEGs; (I) the PPI network of the 54 immune-related DEGs of M0 macrophage cells.; MCODE: Molecular Complex Detection; PPI, protein–protein interaction; STRING, Search Tool for the Retrieval of Interacting Genes; TF, transcription factor.