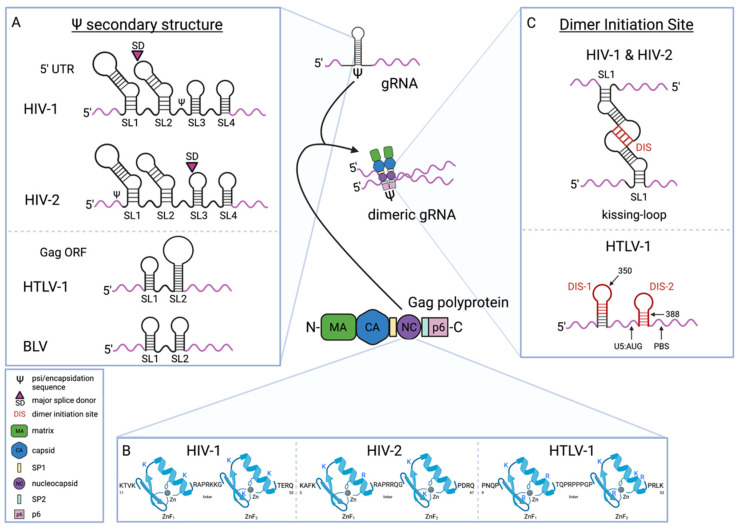
Figure 3.
Schematic representation of the human retroviral gRNA packaging process. Gag stabilization of retroviral genome dimers which occurs between two Gag-gRNA duplexes. (A) HIV-1 and HIV-2 RNA secondary structure models of the 5-untranslated region (5′ UTR) containing the psi (Ψ) sequence with indication of the major splice donor site (SD) either upstream to the Ψ sequences necessary for packaging, as in HIV-1 or downstream to the Ψ packaging sequences as in the case of HIV-2; HTLV-1 and BLV RNA secondary structure models of the gRNA packaging sequence are located in the Gag open reading frame (ORF). The RNA structural elements are marked as indicated: SL1, stem-loop 1; SL2, stem-loop 2; SL3, stem-loop 3; SL4, stem-loop 4; gRNA, viral genomic RNA. Stem-loops are not drawn to scale. (B) Model of the structural motifs in the retroviral Gag polyprotein nucleocapsid (NC) domain mediating the specific Gag-gRNA interaction. Shown are the two zinc-finger (ZnF1 and ZnF2) motifs in HIV-1, HIV-2, and HTLV-1 NC (amino acid residues 11–53, 5–47, and 9–53 respectively). During HIV-1 gRNA packaging, both ZnFs are necessary as the predominant interacting partners with the gRNA, yet the N-terminal ZnF has a greater impact on gRNA packaging. Highlighted in blue are the specific, positively charged, basic amino acid residues within the two ZnF motifs that have been identified to be involved in binding the gRNA. (C) Representative kissing-loop structure induced by NC of HIV-1 and HIV-2 at the gRNA dimer initiation site (DIS), a palindromic sequence located in SL1 of the Ψ sequence secondary structure in the 5′ UTR; representative structure of the two DIS sites identified for HTLV-1 containing palindrome-like sequences located within loop structures in the 5’ leader sequence upstream of the primer binding site (PBS). Created with BioRender.com software (accessed 4 May 2022).