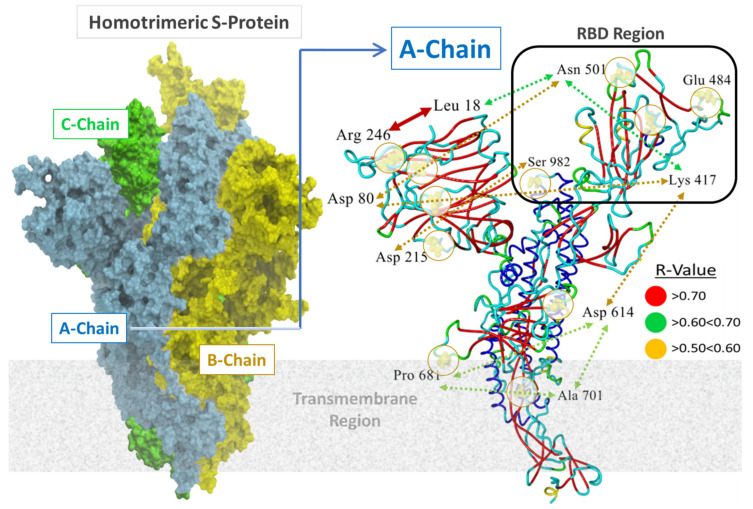
Figure 2.
Left Panel: Surface rendition of the homology construct (Template = PDB 6XR8) of the SARS-CoV-2 S-glycoprotein employed in this study. This full-sequence model (SM#05; residues 14–1146) was constructed and refined in (ICM-Chemist_Pro software; Molsoft, LLC) based on the 3D electron microscopic structure found at PDB 6XR8 (SM ID = 6xr8.1) (see https://swissmodel.expasy.org/repository/species/2697049 for details, accessed on 28 April 2022). Sequence similarity was 0.62. The Global Model Quality Estimation value for this model was 0.73, indicative of a high-quality model. Right Panel: Locations of 11 residues of interest in this study (mutation loci) in the A-chain of Homotrimeric S-Protein are indicated. Double-headed arrows indicate residue pairs that were statistically correlated (R > 0.5) in free-energy protein stability calculations averaged over 20 amino acid substitutions at each locus. Higher correlations indicate mutations at each pair locus affect protein stability to a similar magnitude. For example, sites R246 and L18 exhibited the highest correlation (R = 0.87) suggesting the possibility that mutation-driven conformational changes at one site may influence conformation at the other site in a manner that also favors enhancement of S-protein stability.