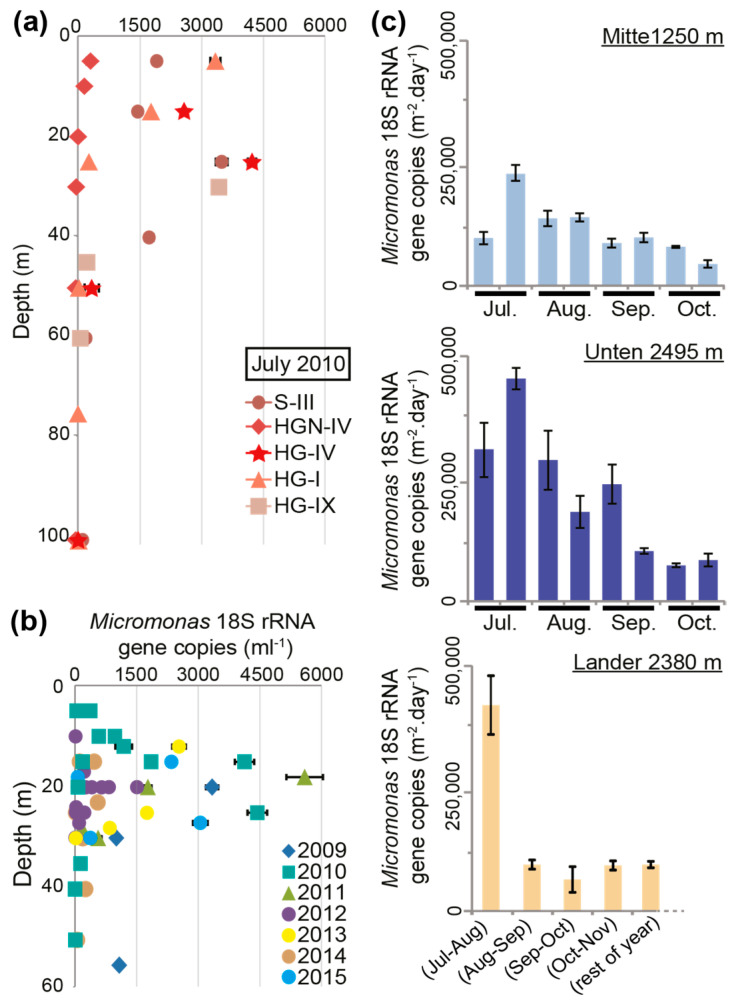
Figure 3.
A multi-year view of Micromonas abundances in the Fram Strait. (a) Micromonas (minimum) 18S rRNA gene copies per mL by qPCR (which, due to losses during extraction, etc., could only capture minimum values) of filtered seawater samples from depth profiles at the five LTER HAUSGARTEN stations sampled by seawater filtration in the month of July, 2010. DNA surface samples are missing above 15 m at HG-IV and above 30 m at HG-IX. (b) Micromonas (minimum) 18S rRNA gene copies per mL from stations along the Svalbard–Greenland transect over a seven-year period that were collected at the subsurface chlorophyll maximum, as defined based on the in vivo chlorophyll a maximum. (c) Deposition (or detectable remainders) of Micromonas in the moored long-term sediment traps and the bottom lander positioned at HAUSGARTEN site HG-IV in 2010. Error bars reflect the standard deviation of technically triplicated qPCR measurements.