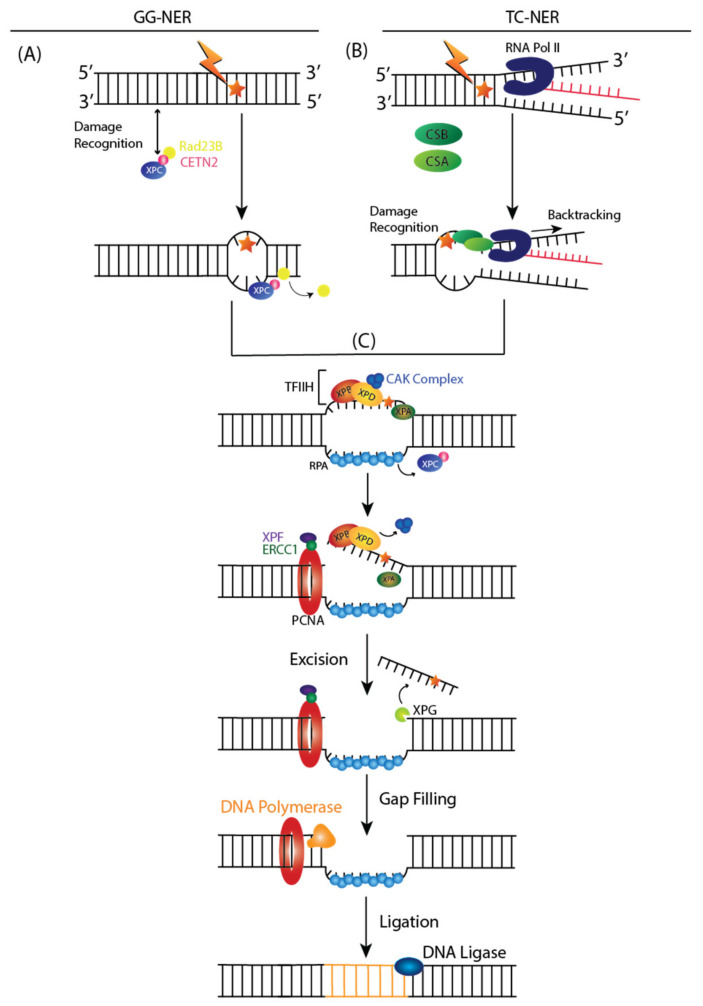
Figure 2.
Nucleotide Excision Repair (NER) Subpathways: (A) Global Genome NER (GG-NER) and (B) Transcription-Coupled NER (TC-NER). (A) Helix distorting lesions are recognized with the help of XPC, Rad23B, and Centrin2 (CETN2). After XPC binds to the damage, Rad32B dissociates from the complex. (B) Damage is indirectly recognized by stalled RNA Pol II at the site of the lesion. CSB-CSA complex is formed, resulting in RNA Pol II backtracking to increase the accessibility of DNA lesions for repair. (C) In both GG-NER and TC-NER, the TFIIH complex is recruited post-lesion recognition. Upon binding of TFIIH, the CAK complex will dissociate from the core of the TFIIH, and the double helix further opens up around the lesion. XPD, XPA, and XPB will verify the existence of the lesions and binds to the single-stranded chemically altered nucleotide. RPA then coats the undamaged strand while XPF-ERCC1 heterodimer will create a 5′ incision to the lesion. Then, XPG is activated and will cut the damaged strand 3′ to the lesion, which excises the strand. PCNA will recruit DNA polymerases for gap-filling DNA synthesis and is finally sealed by DNA Ligase 1 or 3. The orange star denotes DNA damage.