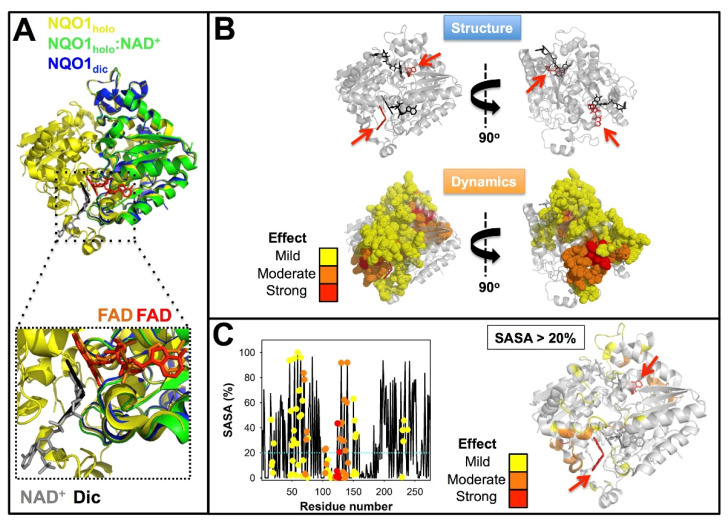
Figure 6.
Structure and dynamics of NQO1 upon binding different ligands. (A) Structural overlay of the X-ray structures of NQO1holo (1D4A) [81], NQO1holo:NAD+ (kindly supplied by Profs. Mario Bianchet and Mario Amzel, John Hopkins University Medical School, Baltimore, Maryland, USA) and NQO1dic (2F1O). The lower panel shows a zoom highlighting the position of the FAD (orange, NQO1dic and red, NQO1holo:NAD+), NAD+ (in grey) and dicoumarol (Dic, in black). (B) Dicoumarol binding causes long-range effects on the structural dynamics of NQO1 WT. Residues shown in dot representation are those for which the structural dynamics is reduced according to HDXMS [132]. (C) Most of the residues whose dynamics are reduced upon dicoumarol binding are solvent-exposed. The plot in the left shows the solvent accessible surface area (SASA) for the each residue as calculated in [132] and color circles indicate the magnitude of the change in structural dynamics. The figure on the right shows the structural location of solvent-exposed residues (SASA > 20%). The color scales in panels B and C reflect the magnitude of the changes in protein dynamics according to [132] and red arrows indicate the position of dicoumarol.