Abstract
The Columbia River estuary is a dynamic system in which estuarine turbidity maxima trap and extend the residence time of particles and particle-attached bacteria over those of the water and free-living bacteria. Particle-attached bacteria dominate bacterial activity in the estuary and are an important part of the estuarine food web. PCR-amplified 16S rRNA genes from particle-attached and free-living bacteria in the Columbia River, its estuary, and the adjacent coastal ocean were cloned, and 239 partial sequences were determined. A wide diversity was observed at the species level within at least six different bacterial phyla, including most subphyla of the class Proteobacteria. In the estuary, most particle-attached bacterial clones (75%) were related to members of the genus Cytophaga or of the α, γ, or δ subclass of the class Proteobacteria. These same clones, however, were rare in or absent from either the particle-attached or the free-living bacterial communities of the river and the coastal ocean. In contrast, about half (48%) of the free-living estuarine bacterial clones were similar to clones from the river or the coastal ocean. These free-living bacteria were related to groups of cosmopolitan freshwater bacteria (β-proteobacteria, gram-positive bacteria, and Verrucomicrobium spp.) and groups of marine organisms (gram-positive bacteria and α-proteobacteria [SAR11 and Rhodobacter spp.]). These results suggest that rapidly growing particle-attached bacteria develop into a uniquely adapted estuarine community and that free-living estuarine bacteria are similar to members of the river and the coastal ocean microbial communities. The high degree of diversity in the estuary is the result of the mixing of bacterial communities from the river, estuary, and coastal ocean.
The degree of bacterial diversity in estuarine environments is expected to be high due to a combination of the mixing of seawater and freshwater and the resuspension of sediments and particles from many sources, including benthic zones, tidal mudflats, and sea grass beds. However, only a fraction of these bacteria may be active as consumers of detrital organic matter. Previous work in the Columbia River estuary showed that the fraction of bacteria attached to particles accounted for approximately 90% of the heterotrophic bacterial activity in the water column and that these bacteria were 10 to 100 times more active than free-living bacteria (5, 6). Particle-attached bacteria are responsible for most of the degradation of detrital organic matter in the estuary (6) and are also part of a thriving estuarine food web in which they are consumed by detritivorous copepods, the dominant metazoan grazers in the system (38). This food web is supported by allochthonous organic material and river phytoplankton, the supply of which far surpasses in situ primary production (39). In the estuary, this material forms organic-rich particles (33) that can be heavily colonized by bacteria and which are the site of the majority of water column extracellular enzymatic activity (6).
The physical, chemical, and biological environments of the Columbia River estuary are centered in the estuarine turbidity maxima (ETM), which are common, well-studied features of river-dominated estuaries, created by the interaction between river flow and tidal forcing (4). In the Columbia River estuary, ETM trap and extend the residence time of particles in the deeper regions of the two main channels near the head of the salt wedge. Organic and inorganic material from the river and the coastal ocean enters into these ETM regions and becomes part of intertidal cycles of sedimentation and resuspension as it is advected up and down the estuary. The residence time of particles in the ETM is thought to be approximately 2 to 4 weeks (35), which is much longer than the 1- to 2-day residence time of water (30). The organic matter associated with these particles is thought to be the primary food source for the food web of the Columbia River estuary (3, 6).
One goal of our research was to investigate whether the estuarine hydrodynamics involved in ETM formation influences the composition of bacterial communities. We hypothesized that actively growing particle-attached bacteria trapped in the ETM form a community that develops and is adapted to life in the estuary and is different from source communities in the river and the coastal ocean. We also hypothesized that the free-living bacterial community in the estuary grows too slowly to develop into an estuarine community and is therefore composed of bacteria from the river and the coastal ocean.
Microbial community analyses using 16S rRNA sequencing have provided a picture of bacterial diversity in oceans, lakes, soils, sediments, aquifers, animal guts, terrestrial hot springs, and sewage (7, 8, 10, 12, 17, 19, 24, 31, 32, 34, 40, 42, 45). No such studies, however, have been conducted on planktonic bacteria in rivers or estuaries. Here we present the results of community analyses of particle-attached and free-living bacteria in the Columbia River of the United States Northwest, its estuary, and the adjacent coastal ocean.
MATERIALS AND METHODS
Sampling.
The Columbia River is the second-largest river in the United States, with a drainage basin of 660,480 km2 (37). Impoundments occur along almost the entire length of the river, creating relatively still reservoirs where riverborne detritus sediments and phytoplankton thrive. The river drains into a shallow, partially mixed estuary (Fig. 1) with two main channels that are generally 20 to 25 m deep. The South Channel is dredged for navigation. Sediments in the main stem of the estuary are sand. The estuary is flanked by tidal mudflats in a few shallow peripheral bays. Salinity intrudes to the ETM regions in the North and South Channels with every tide and can extend up to approximately 20 km from the mouth of the estuary, depending on the river flow and the tide stage.
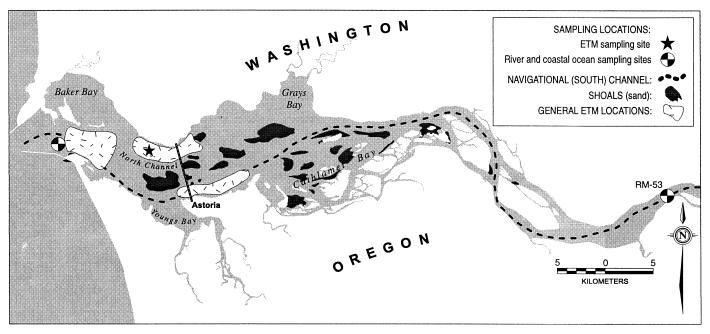
FIG. 1.
The Columbia River estuary with sampling sites.
Water samples were collected at three stations (Fig. 1) in May 1997 with a high-volume, low-pressure pump system coupled to a conductivity-temperature-depth sensor and an optical backscatter sensor for detecting turbidity. The coastal ocean sample was collected about 1 m above the bed at the end of a flood tide in order to collect high-salinity (salinity [S] = 30 psu), low-turbidity (suspended particulate mass [SPM] = 18.5 mg/liter) marine water as it entered the mouth of the estuary. The freshwater river sample was collected at mid-depth (10 m) at a location above the influence of salinity (S = 0 psu, SPM = 29.8 mg/liter). The estuarine sample was collected at an intermediate salinity (S = 9 psu) in the North Channel of the estuary, about 1 m above the bed (17.6 m), during a flood tide turbidity maximum resuspension event in order to obtain ETM particles (SPM = 167.5 mg/liter).
Samples were stored at 4°C for up to 1 h before being processed and then prescreened with a 10-μm-pore-size Nytex mesh to exclude diatom chains and mesozooplankton. Free-living bacteria were gently separated by floating three plastic filter towers (47-mm diameter; Millipore) equipped with 3-μm-pore-size polycarbonate filters (Poretics) on the surface of a sample contained in a 2-liter beaker (6). Filtered water flowed up into the towers and was collected. This method allowed larger particles to settle to the bottom of the beaker, precluding their interference with the filtration of the sample by clogging the filter. Particle-attached bacteria were collected separately by vacuum filtration, again using 3-μm-pore-size polycarbonate filters. Samples were poured into a plastic filter tower and drawn down onto the filters. During intervals when flow through the filters slowed, particles were rinsed by drawing approximately 2 ml of sterile double-distilled water through the filter. Particles were then dislodged from the filter with a stream of sterile water from a squirt bottle, poured out of the filter tower, and collected. Particle-attached and free-living bacteria were then concentrated onto separate 0.2-μm-pore-size Sterivex filters (Millipore). The coastal ocean particle-attached sample was lost during processing, and so a whole-water sample was used instead.
DNA extraction and purification.
Sterivex filters were immediately flooded with approximately 2 ml of sarcosyl lysis buffer (0.14 M NaCl; 50 μM sodium acetate [pH 5.2]; 0.3% N-lauroylsarcosine, sodium salt [sarcosyl]; autoclaved and filter sterilized) and 100 μl of a 2% proteinase K solution, agitated briefly, and incubated at 38°C for 2 h. The filters were then frozen at −20°C until further processed.
The filters were defrosted and agitated, and each sample was drawn off along with resuspended particulate material. Particles carried off the filter with the extraction buffer were pelleted by centrifugation (6,000 × g, 5 min). This was done for all samples. The resulting supernatant was combined with 0.5 ml of a 5% cetyltrimethylammonium bromide (CTAB) solution and 20 μl of a 20% proteinase K solution, incubated at 37°C on a rotating carousel for 30 min, and stored on ice.
DNA extraction buffer (0.1 M Tris-HCl [pH 8], 0.1 M Na-EDTA [pH 8], 0.1 M Na2H2PO4 [pH 8], 1.5 M NaCl, 5% CTAB) (46) and proteinase K (2%) were added to both parts of each sample: the Sterivex filter (1.85 ml and 10 μl, respectively) and pelleted particles (0.925 ml and 5 μl, respectively). Samples were frozen (at −70°C) and thawed (at 65°C) three times and then incubated on a rotating carousel at 37°C for 30 min. Sodium dodecyl sulfate (SDS; 20%) was added (150 μl to the filter; 30 μl to the particles), and samples were incubated on a rotating carousel at 65°C for 2 h. The particles were centrifuged, and the buffer was added to the supernatant from the first extraction. The extraction buffer from the Sterivex filter was then centrifuged over the particles, and the supernatant was added to the supernatant from the first extraction. The extraction procedure was repeated once on the Sterivex filter and the particles.
An equal volume of chloroform-isoamyl alcohol (24:1) was added to the combined supernatants, and the solution was vortexed and centrifuged (3,000 rpm in a Jouan microcentrifuge; 10 min). The aqueous layer was transferred to a sterile 50-ml glass Corex tube (Corning), combined with an equal volume of isopropanol, and incubated at room temperature for 1 h. Precipitated DNA was centrifuged (16,000 × g, 20 min, room temperature), washed with 5 ml of 70% ethanol, dried down, dissolved in 500 μl of TE buffer (10 mM Tris-HCl, 1 mM Na-EDTA; pH 8), and frozen.
Aliquots of each DNA extract were purified with Qiaquick PCR purification columns (Qiagen) in accordance with the manufacturer’s instructions except that the DNA was washed twice with PE buffer and eluted twice with EB buffer heated to 65°C (buffers were provided by the manufacturer).
Clone library construction.
PCR was performed on 8 to 10 separate 100-μl reaction mixtures (2.5 mM MgCl, 0.8 mM deoxynucleoside triphosphates, 1 ng of each primer/μl, 2.5 U of Taq DNA polymerase [Promega], 1× PCR buffer [Promega]) for each DNA sample, using universal bacterial primer 8f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and universal primer 1492r (5′-GGT TAC CTT GTT ACG ACT T-3′). PCR amplification began with a 1-min denaturation at 94°C; this was followed by 20 to 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min. The final cycle was extended at 72°C for 5 min. PCR cycles were stopped while the product concentration was still increasing exponentially. The resulting low concentrations of PCR product required us to run multiple PCRs to have enough product for cloning. PCR products were combined, concentrated, and purified with Qiaquick PCR purification columns (Qiagen) in accordance with the manufacturer’s instructions.
PCR products were ligated into the pGEM-T cloning vector (Promega) and used to transform JM109 competent cells (Promega) as per the manufacturer’s instructions. Positive colonies were picked, stored on agar plates, and frozen in a liquid medium at −70°C.
The use of environmental clone libraries as a quantitative measure of diversity has fallen into question due to variations in primer specificity and overamplification of rare sequences (15, 41, 44). However, 16S rRNA clone libraries have provided valuable qualitative pictures of microbial diversity that allow us to compare and contrast the communities in different environments (10, 26, 29, 47). We attempted to minimize the amplification of contaminant genes and the overamplification of rare genes by using a reduced number of PCR cycles, stopping the amplification while the concentration of PCR product was still increasing exponentially.
Restriction fragment length polymorphism (RFLP) analysis.
Seventy-five clones from each of the two estuarine clone libraries were randomly chosen to inoculate 100-μl aliquots of Luria-Bertani broth medium (1% Tryptone, 0.5% yeast extract, 1% NaCl; pH 7), with incubation at 37°C for 1 h. The plasmid inserts were PCR amplified with the vector-specific primers SP6 (5′-ATT TAG GTG ACA CTA TAG-3′) and T7 (5′-TAA TAC GAC TCA CTA TAG GG-3′) (20-μl reaction volumes with 1 μl of clone culture, 3 mM MgCl, 0.8 mM deoxynucleoside triphosphates, 1 ng of each primer/ml, 2.5 U of Taq DNA polymerase [Promega], and 1× PCR buffer [Promega]). PCR amplification involved a 1-min denaturation at 94°C followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min, after which was performed a 5-min extension at 72°C. PCR products were restriction digested with MspI and RsaI (Boehringer-Mannheim) in accordance with the manufacturer’s instructions, electrophoresed on 2.5% agarose gels (Agarose 3:1; Amresco) prepared with TAE (0.04 M Tris-acetate, 1 mM EDTA), and stained with SYBR green (Molecular Probes). Gel images were digitized with a Fluorimager 575 fluorescent gel scanner (Molecular Dynamics), and band sizes were determined by using the program FragmeNT Analysis version 1.1 (Molecular Dynamics) based on a 1-kb ladder size standard (Gibco BRL).
Sequencing.
Estuarine clone inserts with unique RFLP patterns and 25 clones chosen randomly from each of the river and coastal ocean clone libraries were sequenced. Inserts were PCR amplified as described above. PCR products were purified by using Qiaquick PCR purification columns, sequenced bidirectionally with primers 8f (see above) and 519r (5′-GWA TTA CCG CGG CKG CTG-3′), and a PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems Inc.), and resolved on a model 373A automated DNA sequencer (Applied Biosystems Inc.).
Phylogenetic analysis.
We tested for chimeric sequences in two ways. First we ran sequences through the Chimera Check program of the Ribosomal Database Project (RDP) website (25). We also analyzed the secondary structures by aligning bases 34 to 40 with bases 763 to 755 and bases 304 to 333 with bases 524 to 560 (Escherichia coli numbering system) for each sequence (22).
Unaligned sequences were submitted to the Sequence Match program of the RDP and to the Advanced BLAST search program of the National Center for Biotechnology Information (NCBI) website (29a) to find closely related sequences. Related sequences were acquired by using the Batch Entrez program (NCBI). Preliminary alignments were made by using the Sequence Align program (RDP), requesting that common gaps be preserved. Sequences were organized by phylum, and alignments were completed manually by using the SeqApp program (11a). Some sections of the sequences in each phyla could not be aligned and were therefore not used in subsequent analyses (Table 1).
TABLE 1.
Bases used for phylogenetic analyses (E. coli numbering system)
| Major grouping | Range of bases used for analyses | Bases omitted |
|---|---|---|
| α-Proteobacteria | 32–479 | 71–98 |
| β-Proteobacteria | 33–514 | 76–93 |
| γ-Proteobacteria | 50–494 | 76–93, 199–220 |
| δ-Proteobacteria | 28–519 | 76–93, 184–193 |
| Gram-positive bacteria | 37–508 | 71–97, 450–479 |
| Cytophaga-Flavobacteria | 73–513 | None |
| Verrucomicrobiales and Planctomyces | 37–519 | 71–97, 184–193, 453–477 |
| Cyanobacteria and chloroplasts | 94–451 | None |
| Unknown | 50–519 | 69–99, 184–220, 452–480 |
Percent similarity between sequences was determined by using the Distances program of the Wisconsin Package version 9.1-UNIX of the Genetics Computer Group, Inc., set to calculate uncorrected distances.
Phylogenetic analysis was accomplished with the PAUP program (Smithsonian Institution, 1997) accessed through the Wisconsin Package. Consensus (50% majority rule) trees were constructed by using uncorrected neighbor-joining distances with 1,000 bootstrap replicates. These trees exclude groupings that occurred in less than 50% of the replicates. Negative branch lengths were prohibited.
Phylum-specific trees were originally prepared with three different outgroup sequences from among the following organisms: Roseobacter denitrificans, Rhodoferax fermentans, E. coli, Pirellula staleyi, Cytophaga lytica, and Agrococcus jenensis. Clones were placed on trees with their closest relatives identified by the database searches described above. Clones with no clear affiliation to a single phylum were put on a separate tree with a broad diversity of bacteria, using an archaebacterium as an outgroup.
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers for sequences determined in these studies are as follows: for CR-FL1 to -6, -8 to -13, -15, -16, -18, -20 to -23, and -25, -30, AF141387 to AF141411; for CR-PA2, -6, -11, -13, -15, -16, -19 to -22, -24, -26, -27, -30, -36, -38, -40, -43, -44, -50, -52, -53, and -55, AF141412 to AF141434; for CRE-FL1, -3, -4, -7, -8, -10, -11, -13, -14, -16, -18 to -26, -28, -31, -33, -35, -37 to -41, -43 to -47, -49, -50, -52 to -54, -56, -57, -59 to -64, -67 to -70, and -72 to -80, AF141435 to AF141493; for CRE-PA2, -4, -6, -7, -9 to -11, -14 to -18, -21 to -27, -29, -30, -32, -34, -35, -37 to -42, -44, -45, -47, -49, -50 to -53, -58 to -60, -63, -64, -66, -69, -70, -72 to -80, and -82 to -89, AF41494 to AF141556; for CRO-1, -2, -4, -6, -11, -13 to -19, -21, -22, -24, -27 to -29, and -31 to -35, AF141557 to AF141579; and for CRO-FL1 to -5, -7 to -18, and -22 to -26, AF141580 to AF141601.
RESULTS
Heterotrophic bacterial activities ([3H]thymidine incorporation rate) determined within 1 week of DNA sampling, 1 year, and 2 years prior to this study (6) are summarized in Table 2. Estuary bacterial cell concentrations in the particle-attached and free-living fractions were similar, but the activity associated with the particle-attached bacteria was always higher than that associated with the free-living bacteria. Also, the overall bacterial activity in the estuary was higher than that in the coastal ocean or the river.
TABLE 2.
Bacterial abundance and thymidine incorporation rates in samples collected in the spring from the Columbia River, estuary, and adjacent coastal ocean
| Sample location and site | Bacteria abundance (109 liter−1)
|
Thymidine incorporation rate (pmol liter−1 h−1)
|
||||
|---|---|---|---|---|---|---|
| Mean | Range | n | Mean | Range | n | |
| Columbia River | ||||||
| 1997 | ||||||
| Free-living | 0.4 | 0.2–1.0 | 4 | |||
| Particle attached | 1.3 | 0.9–2.1 | 4 | |||
| 1996 | ||||||
| Free-living | 0.9 | 0.8–1.0 | 2 | |||
| Particle attached | 13.7 | 11.5–15.8 | 2 | |||
| 1995 | ||||||
| Free-living | 0.3 | 0.3–0.3 | 2 | |||
| Particle attached | 2.2 | 2.0–2.5 | 2 | |||
| Columbia River estuary | ||||||
| 1997 | ||||||
| Free-living | 1.8 | 0.9–3.8 | 10 | 1.0 | 0.1–2.6 | 35 |
| Particle attached | 2.4 | 0.7–5.1 | 10 | 7.7 | 0.3–31.7 | 35 |
| 1996 | ||||||
| Free-living | 8.9 | 0.2–37.3 | 20 | |||
| Particle attached | 57.4 | 12.8–104.4 | 20 | |||
| 1995 | ||||||
| Free-living | 1.4 | 0.7–2.6 | 56 | 3.1 | 0.2–7.7 | 54 |
| Particle attached | 3.3 | 0.1–12.6 | 56 | 38.4 | 12.0–90.1 | 54 |
| Coastal ocean, 1997 | ||||||
| Free-living | 0.5 | 0.2–1.0 | 2 | |||
| Particle attached | 1.3 | 1.2–1.7 | 2 | |||
RFLP patterns were used as an initial measure of diversity in the estuarine clone libraries. The goal was to sequence representatives from groups of clones with identical RFLP patterns. However, only 43 of 146 clones examined were part of groups with matching RFLP patterns, and 25 of the 43 were in one group. Representatives from three groups of clones with matching RFLP patterns were sequenced and were found to be identical (CRE-FL4 and -7, CRE-PA2 and -16, and CRE-FL16 and -19). Based on this evidence, 24 estuarine clones were categorized by their RFLP patterns and were not sequenced (Table 3; Fig. 2).
TABLE 3.
Clone sequences from each clone library, listed with phylum affiliation, nearest neighbor from the global database, percent similarity based on alignable base pairs, and grouping within each phylum
| Sample location and type | Category | Clone no.a | Nearest neighbor | % Similarity | Assemblage |
|---|---|---|---|---|---|
| Coastal ocean, free-living | α-Proteobacteria | CRO-FL4, -10, -15, -23 | OCS12 | 97.6–99.7 | SAR11 |
| CRO-FL1, -3, -7, -12, -26 | OCS154 | 96.3–98.4 | SAR11 | ||
| CRO-FL11 | OCS124 | 99.7 | OCS124 | ||
| CRO-FL5, -16 | ?d | ? | ? | ||
| β-Proteobacteria | CRO-FL2 | BAL47 | 98.1 | Rubrivivax spp. | |
| CRO-FL17 | Hydrogenophaga flava | 93.7 | Rubrivivax spp. | ||
| CRO-FL25 | ? | ? | ? | ||
| γ-Proteobacteria | CRO-FL8 | NKB4 | 90.5 | ||
| Gram-positive bacteria | CRO-FL9, -14 | ACK-M1 | 94.2, 94.9 | ACK-4 | |
| CRO-FL22 | OCS155 | 99.8 | OM1 | ||
| Cyanobacteria | CRO-FL18, -24 | Prochlorococcus sp. strain MIT9303 | 98.6, 97.8 | Prochlorococcus spp. | |
| CRO-FL13 | SAR7 | 99.5 | SAR7 | ||
| Coastal ocean, particle-attached | α-Proteobacteria | CRO1 | OM42 | 98.9 | Marine Rhodobacter spp. |
| γ-Proteobacteria | CRO2, -21 | Legionella lytica | 95.1, 94.7 | Legionella spp. | |
| CRO33 | Legionella feeleii | 94.7 | Legionella spp. | ||
| CRO14, -19 | Pseudomonas syringae | 99.4 | Pseudomonas spp. | ||
| CFBb | CRO4 | MED25 | 92.2 | Cytophaga spp. | |
| Planctomyces spp. | CRO13 | Planctomyces limnophilus | 98.5 | Planctomyces limnophilus | |
| Cyanobacteria | CRO15, -34 | Prochlorococcus sp. strain MIT9303 | 98.1, 97.3 | Prochlorococcus spp. | |
| CRO16, -24, -27, -29, -31, -35 | SAR7 | 98.9–99.7 | SAR7 | ||
| Chloroplasts | CRO11, -22, -28, -32 | OM81 | 91.0 | Chrysophyceae | |
| CRO17 | OM81 | 87.5–88.0 | Chrysophyceae | ||
| Unknown | CRO6 | ? | ? | ? | |
| CRO18 | ? | ? | ? | ||
| Columbia River, free-living | α-Proteobacteria | CR-FL10 | LD12 | 100.0 | Freshwater SAR11 |
| CR-FL11 | Soil clone (AF010012) | 96.6 | Rhizobium-Agrobacterium | ||
| β-Proteobacteria | CR-FL2, -6, -9 | BAL47 | 96.3–96.5 | Rubrivivax spp. | |
| CR-FL8* | MT11 | 94.6 | Rubrivivax spp. | ||
| CR-FL13, -22 | LD17 | 97.0, 96.7 | Polynucleobacter necessarius | ||
| CR-FL23 | ACK-L6 | 96.1 | Polynucleobacter necessarius | ||
| CR-FL21 | ACK-C30 | 99.8 | Methylophilus spp. | ||
| γ-Proteobacteria | CR-FL28 | Vibrio marinus | 92.9 | ? | |
| CR-FL29 | Pseudomonas sp. clone (U63942) | 95.8 | Pseudomonas spp. | ||
| Gram-positive bacteria | CR-FL16, -18 | MC19 | 84.2, 86.3 | CR-FL16 | |
| CR-FL3, -20 | ACK-M1 | 88.6, 91.1 | ACK-4 | ||
| CR-FL30 | Agrococcus jenensis | 86.8 | ? | ||
| CFB | CR-FL26 | Capnocytophaga gingivalis | 85.4 | Cytophaga spp. | |
| CR-FL12 | ? | ? | ? | ||
| Planctomyces spp. | CR-FL15 | MC55 | 88.7 | Isophaera spp. | |
| Verrucomicrobiales spp. | CR-FL1, -25, -27 | VeSm13 | 86.8–87.6 | Verrucomicrobiales | |
| CR-FL5 | MC18 | 93.3 | Verrucomicrobiales | ||
| Unknown | CR-FL4 | ? | ? | ? | |
| Columbia River, particle-attached | α-Proteobacteria | CR-PA55 | Rhodobacter sphaeroides | 94.2 | Freshwater Rhodobacter spp. |
| CR-PA22 | Beijerinckia indica | 95.3 | Rhizobium-Agrobacterium spp. | ||
| CR-PA53 | MHP17 | 93.9 | Rhizobium-Agrobacterium spp. | ||
| β-Proteobacteria | CR-PA6, -11 | Rhodoferax fermentans | 98.3, 96.3 | Rubrivivax spp. | |
| CR-PA24 | Alcaligenes denitrificans | 92.5 | Bordetella spp. | ||
| CR-PA50 | Ralstonia pickettii | 99.3 | Ralstonia spp. | ||
| γ-Proteobacteria | CR-PA40 | Methylobacter sp. strain BB5.1 | 96.2 | Methylomonas spp. | |
| CR-PA44 | Legionella feeleii | 96.0 | Legionella spp. | ||
| CR-PA27 | TRS20 | 88.2 | ? | ||
| Gram-positive bacteria | CR-PA52 | MC19 | 86.5 | CR-FL16 | |
| CR-PA21, -26, -38 | MC19 | 86.1–87.3 | CR-FL16 | ||
| CR-PA36 | ACK-M1 | 94.7 | ACK-4 | ||
| CR-PA13 | OPB90 | 83.0 | ? | ||
| CFB | CR-PA19 | Soil clone C125 (AF013539) | 93.4 | Saprospira spp. | |
| Planctomyces spp. | CR-PA16 | MC55 | 88.7 | Isophaera spp. | |
| Chloroplasts | CR-PA2, -20 | Chloroplast (Skeletonema pseudocostatum) | 98.6, 98.4 | Bacillariophyta | |
| CR-PA30, -43 | Hstp14 | 98.1, 97.8 | Bacillariophyta | ||
| Unknown | CR-PA15 | ? | ? | ? | |
| Columbia River estuary, free-living | α-Proteobacteria | CRE-FL64 | OM42 | 99.5 | Marine Rhodobacter spp. |
| CRE-FL23 | MED26 | 97.4 | Marine Rhodobacter spp. | ||
| CRE-FL63 | OCS12 | 98.7 | SAR11 | ||
| CRE-FL21 | Sphingomonas adhaesiva | 95.3 | Sphingomonas spp. | ||
| CRE-FL20 | ? | ? | |||
| CRE-FL1 | ? | ? | |||
| β-Proteobacteria | CRE-FL38, -49 | BAL47 | 98.1 | Rubrivivax spp. | |
| CRE-FL16, -19 (-2, -65) | BAL47 | 95.5 | Rubrivivax spp. | ||
| CRE-FL37c | MT11 | 95.0 | Rubrivivax spp. | ||
| CRE-FL62 | Aquaspirillum delicatum | 92.0 | Rubrivivax spp. | ||
| CRE-FL35, -41, -50 | Rhodoferax fermentans | 96.1–97.2 | Rubrivivax spp. | ||
| CRE-FL14, -26, -79 | ACK-L5 | 98.9–99.6 | Polynucleobacter necessarius | ||
| CRE-FL11 | LD17 | 97.0 | Polynucleobacter necessarius | ||
| CRE-FL45, -78 | ACK-L6 | 96.5, 96.7 | Polynucleobacter necessarius | ||
| CRE-FL73 | ACK-C30 | 100.0 | Methylophilus spp. | ||
| (CRE-FL15) | ACK-C30 | (98.1) | Methylophilus spp. | ||
| CRE-FL40 | ACK-C30 | 95.0 | Methylophilus spp. | ||
| CRE-FL44, -56 (-58) | Alcaligenes denitrificans | 93.1, 92.2 | Bordetella spp. | ||
| CRE-FL33 | Ralstonia pickettii | 98.9 | Ralstonia spp. | ||
| CRE-FL22 | Ultramicrobacterium sp. strain ND5 | 95.7 | ? | ||
| CRE-FL68 | Gallionella ferruginea | 95.5 | Gallionella spp. | ||
| γ-Proteobacteria | CRE-FL4, -7, -61, -76, -77, -80, (-6, -9, -29, -30, -32, -34, -36, -42) | Marinomonas vaga | 89.2–89.8 | Oceanospirillum spp. | |
| CRE-FL8 | Marinomonas aquaeolei | 88.6 | Oceanospirillum spp. | ||
| δ-Proteobacteria | CRE-FL54 | Desulfosarcina variabilis | 93.1 | Desulfobacter spp. | |
| Gram-positive bacteria | CRE-FL67 | MC19 | 84.9 | CR-FL16 | |
| CRE-FL47, -53 | MC19 | 86.8, 85.8 | CR-FL16 | ||
| CRE-FL18, -70 (-66) | ACK-M1 | 90.8, 90.6 | ACK-4 | ||
| CRE-FL13 | ACK-M1 | 88.6 | ACK-4 | ||
| CRE-FL43, -60 | Agrococcus jenensis | 93.3, 94.2 | ? | ||
| CRE-FL10, -72 | OCS155 | 99.5, 99.8 | OM1 | ||
| CFB | CRE-FL46 | Sea ice psychrophile (U85888) | 93.4 | Cytophaga spp. | |
| CRE-FL24, -25 | TBS22 | 96.1, 93.1 | Cytophaga spp. | ||
| CRE-FL57 | OM271 | 94.5 | Cytophaga spp. | ||
| CRE-FL75 | Psychroserpens burtonensis | 88.1 | Cytophaga spp. | ||
| (CRE-FL17) | SCB37 | (93.1) | Cytophaga spp. | ||
| CRE-FL3, -39 | Flectobacillus major | 86.2, 86.4 | Flexibacter flexilis | ||
| Order Verrucomicrobiales | CRE-FL31 | TM18 | 88.5 | Verrucomicrobiales | |
| CRE-FL59 | LD29 | 87.3 | Verrucomicrobiales | ||
| CRE-FL74 | Verrucomicrobium spinosum | 85.0 | Verrucomicrobiales | ||
| Chloroplast | CRE-FL52 | OM20 | 90.5 | Bacillariophyta | |
| Unknown | CRE-FL28 | ? | ? | ? | |
| CRE-FL69 | ? | ? | ? | ||
| Columbia River estuary, particle-attached | α-Proteobacteria | CRE-PA76, -77 | KAT10 | 95.0 | Marine Rhodobacter |
| CRE-PA4, -47, -80, -89 | Rhodobacter capsulatus strain ATH | 97.4–98.4 | Freshwater Rhodobacter | ||
| CRE-PA51 | BAL27 | 95.0 | Freshwater Rhodobacter | ||
| CRE-PA70 | OM188 | 100.0 | SAR11 | ||
| CRE-PA52, -53 | Blastobacter natatorius | 97.1 | Sphingomonas spp. | ||
| β-Proteobacteria | CRE-PA69 | Rubrivivax gelatinosus | 93.3 | Rubrivivax spp. | |
| (CRE-PA65) | Rhodoferax fermentans | (96.1) | Rubrivivax spp. | ||
| CRE-PA22 | ? | ? | ? | ||
| CRE-PA84 | ? | ? | ? | ||
| CRE-PA45 | ACK-C30 | 98.1 | Methylophilus spp. | ||
| γ-Proteobacteria | CRE-PA2, -16, -49, -86, -87, -88 (-5, -8, -20, -33, -48) | Marinomonas vaga | 89.2–89.8 | Oceanospirillum spp. | |
| CRE-PA40 | OM23 | 97.9 | Oceanospirillum spp. | ||
| CRE-PA14, -50 | Symbiont (hydrothermal vent mussel) | 92.3, 93.4 | Thiothrix nivea | ||
| CRE-PA25 | NKB4 | 88.2 | ? | ||
| CRE-PA78 | OM60 | 92.8 | ? | ||
| CRE-PA17 | TBS23 | 89.0 | ? | ||
| CRE-PA9 | Methylococcus capsulatus | 87.8 | Methylomonas spp. | ||
| CRE-PA58, -74 | TRS20 | 88.2, 89.4 | ? | ||
| CRE-PA35 | Xanthomonas vesicatoria | 92.2 | Xanthomonas spp. | ||
| δ-Proteobacteria | CRE-PA6, -66 | Desulfurhopalus vacuolatus | 91.8, 93.1 | Desulfobacter spp. | |
| CRE-PA18 | Desulfovibrio sp. strain STL6 | 94.5 | Desulfovibrio spp. | ||
| Gram-positive bacteria | CRE-PA41 | MC19 | 87.3 | CR-FL16 | |
| CRE-PA39 | OCS155 | 99.5 | OM1 | ||
| CRE-PA63 (-67, -81) | OPB90 | 83.0 | ? | ||
| CRE-PA72 | Spiroplasma sp. strain Y32 | 83.2 | Low G+C | ||
| CRE-PA64 | MB2424 | 88.5 | Low G+C | ||
| CFB | CRE-PA32 | BAL13 | 93.6 | Cytophaga spp. | |
| CRE-PA38 | Sea ice psychrophile (U85888) | 93.4 | Cytophaga spp. | ||
| CRE-PA10, -79 (-43) | MED25 | 92.0 | Cytophaga spp. | ||
| CRE-PA44 | MED18 | 92.5 | Cytophaga spp. | ||
| CRE-PA37 | SCB37 | 93.1 | Cytophaga spp. | ||
| CRE-PA11, -15, -85 | Psychroserpens burtonensis | 93.0 | Cytophaga spp. | ||
| CRE-PA7 | Flectobacillus major | 86.7 | Flexibacter flexilis | ||
| CRE-PA83 | Soil clone C125 | 89.8 | Saprospira spp. | ||
| CRE-PA30 | ? | ? | ? | ||
| CRE-PA75 | ? | ? | ? | ||
| CRE-PA82 | ? | ? | ? | ||
| Planctomyces spp. | CRE-PA34 | MC100 | 94.6 | Planctomyces limnophilus | |
| Verrucomicrobiales | CRE-PA23 | LD29 | 87.9 | Verrucomicrobiales | |
| CRE-PA73 | MC18 | 92.2 | Verrucomicrobiales | ||
| CRE-PA29 | ? | ? | Verrucomicrobiales | ||
| Chloroplast | CRE-PA60 | Chloroplast (Skeletonema pseudocostatum) | 98.4 | Bacillariophyta | |
| CRE-PA42 | Hstp14 | 96.5 | Bacillariophyta | ||
| CRE-PA59 (-19) | AGG56 | 97.3 | Bacillariophyta | ||
| CRE-PA21 | OM81 | 87.7 | Chrysophyceae | ||
| CRE-PA27 | Chlorella saccharophila | 91.5 | Green plant chloroplasts | ||
| Unknown | CRE-PA24, -26 | ? | ? | ? |
Clone numbers in parentheses were categorized by RFLP pattern.
CFB, Cytophaga-Flexibacter-Bacteroides.
Clone that is most closely related to known contaminants from a negative-control library (43).
?, nearest neighbor could not be determined.
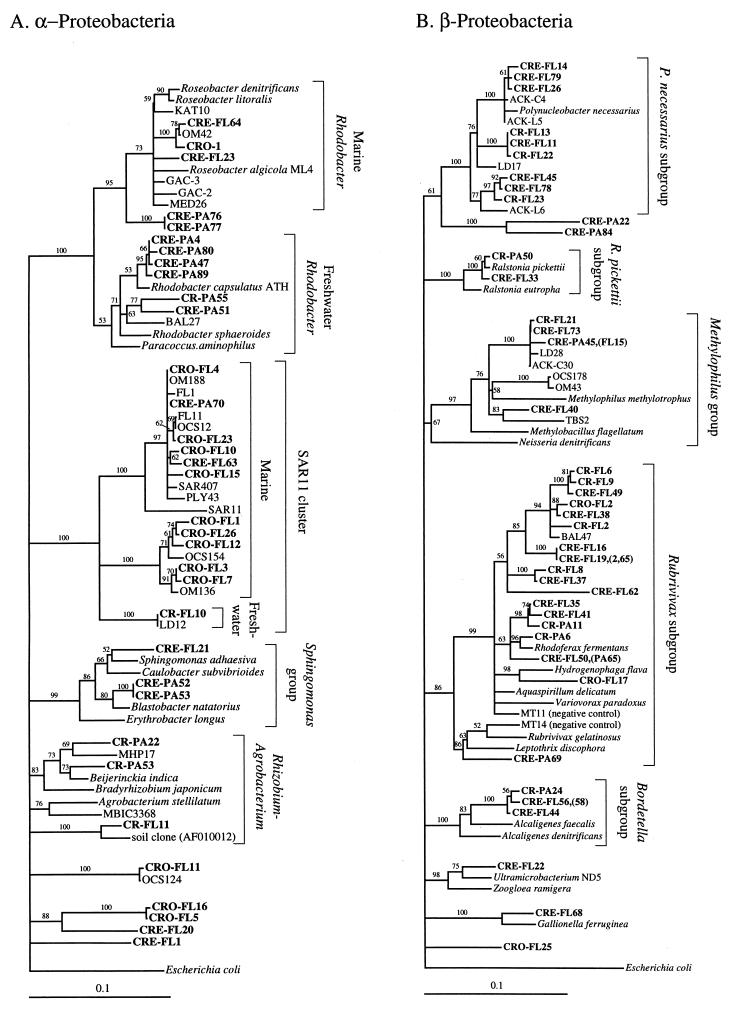
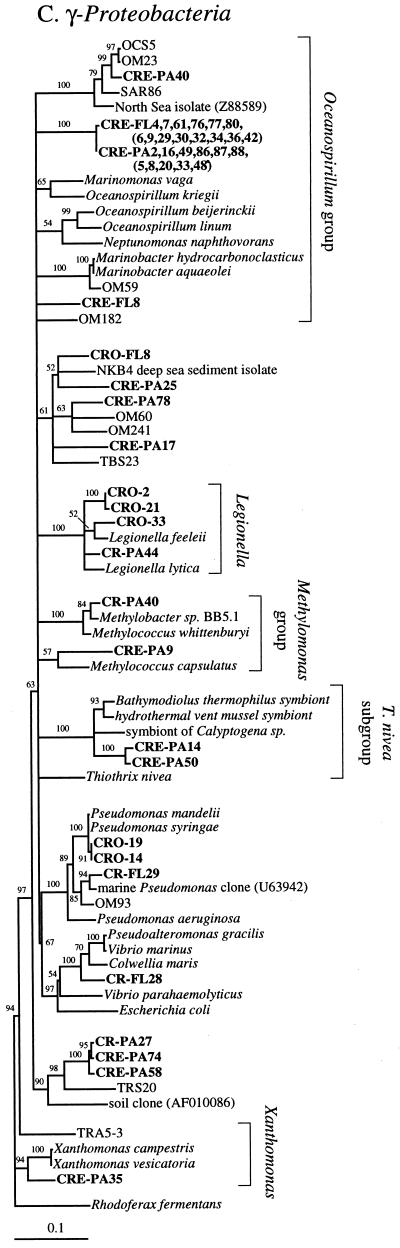
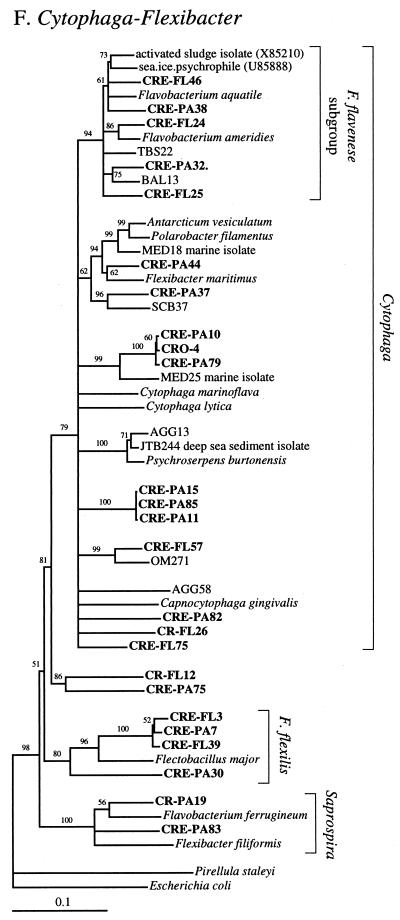
FIG. 2.
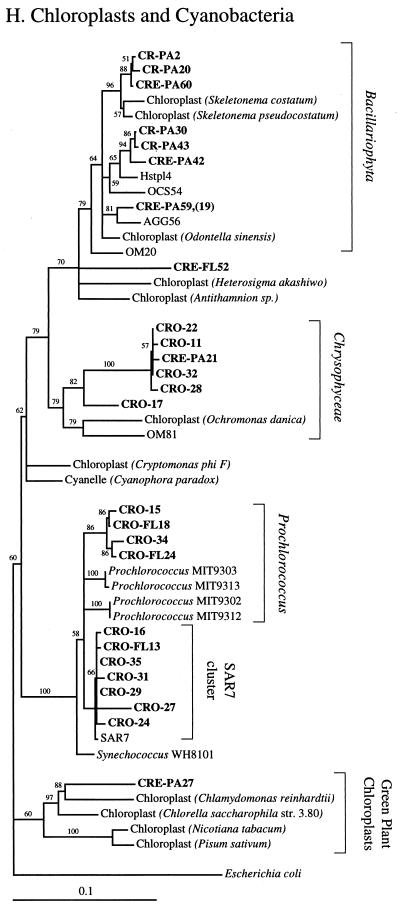
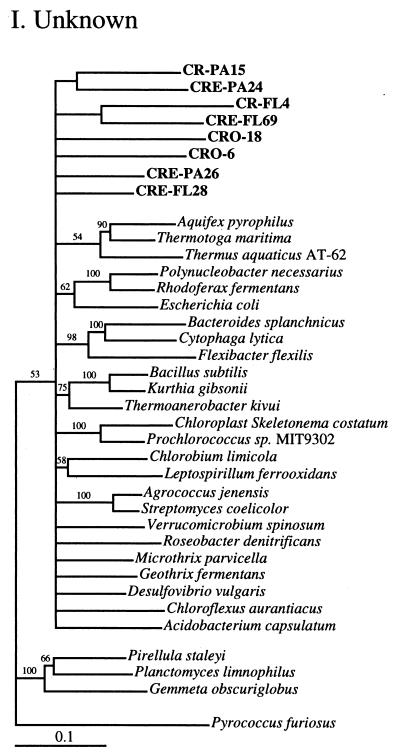
Phylogenetic relationships among 16S rRNA sequences from Columbia River, estuary, and adjacent coastal ocean clones and from other environmental clones and cultured organisms. (A) α-proteobacteria; (B) β-proteobacteria; (C) γ-proteobacteria; (D) δ-proteobacteria; (E) Verrucomicrobiales and Planctomyces clade; (F) Cytophaga-Flexibacter assemblage; (G) gram-positive bacteria; (H) chloroplasts and cyanobacteria; (I) all other clones. Fifty percent majority-rule trees were constructed by the neighbor-joining method. The percentages of 1,000 bootstrap replicates that supported the branching order are shown above or near the relevant nodes. The scale bars correspond to a 10% difference in nucleotide sequence. Clones from this study are indicated in boldface and are named with the following prefixes, designating their sources: CR, Columbia River; CRE, Columbia River estuary; CRO, coastal ocean; PA, particle attached; and FL, free-living. All sequences are available from the GenBank database, and accession numbers are provided if the organism or clone name is not unique. CFB, Cytophaga-Flexibacter-Bacteroides phylum.
All clone sequences from this study are presented on phylum- or subphylum-specific trees (Fig. 2) and listed by clone library (Table 3). Clones with sequences that could not be grouped with known phyla or subphyla were put on a tree with a diverse group of bacteria (Fig. 2I).
Riverine diversity.
This study is the first to investigate the planktonic bacterial community in a river by using 16S rRNA clone libraries. The sequences of 22 of 48 clones from the two river clone libraries were remarkably similar to sequences found in lakes in the Adirondack Mountains, The Netherlands, and Alaska, further confirming the existence of clades of cosmopolitan freshwater bacteria within the α subclass of the class Proteobacteria, the β-proteobacteria, and the order Verrucomicrobiales and among gram-positive organisms (2, 17, 26, 47, 48) (Fig. 2A, B, E, and G). Twelve clones were related to soil isolates and clones, including TRS20 (a γ-Proteobacterium), MC55 (a planctomycete), MC19 (a gram-positive bacterium), and clones related to members of the Rhizobium-Agrobacterium group (α-proteobacteria) (Fig. 2A, C, E, and G). Previously described “freshwater” bacteria clades Rhizobium-Agrobacterium and Verrucomicrobiales also include many isolates and clone sequences from soils. There is probably a close relationship and significant overlap in communities between soil and freshwater bacteria due in part to the interaction between the two environments.
The free-living and particle-attached clone libraries from the river contained clones from the same phyla and subphyla (Fig. 3B), but within these major groupings, free-living and particle-attached clones rarely clustered together. For example, within the β-proteobacteria, clones related to Polynucleobacter necessarius were all free-living, but clones related to BAL47 were all particle attached (Fig. 2B). Chloroplast clones were found only in the particle-attached fraction, probably because their phytoplankton hosts could not pass through the 3-μm-pore-size filter (Fig. 2H). Also, clones related to the order Verrucomicrobiales were found only in the free-living fraction (Fig. 2E).
FIG. 3.
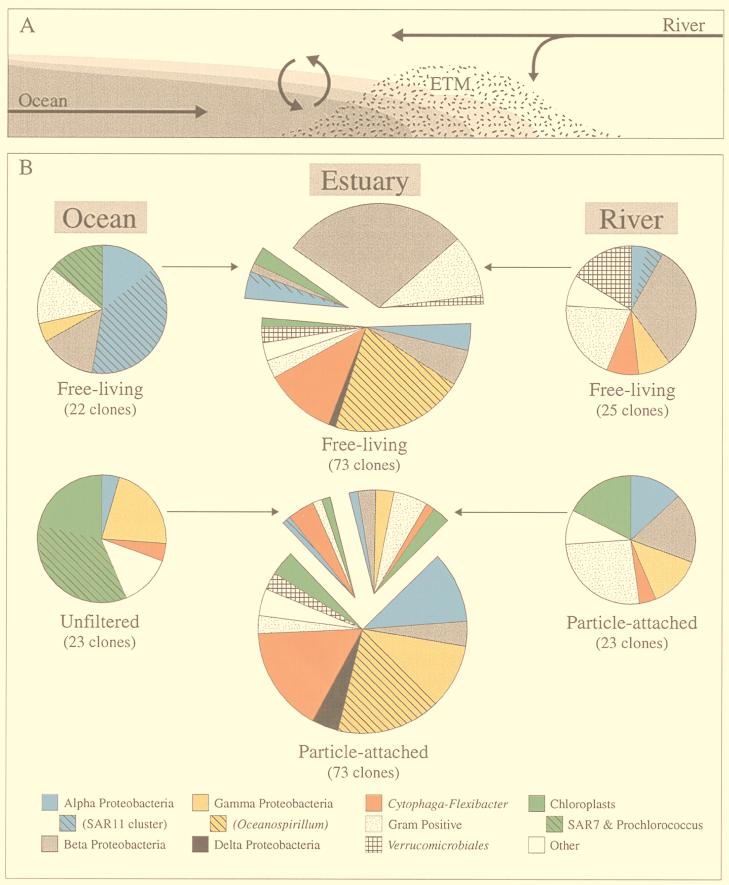
(A) Longitudinal cross section of an ETM region of the Columbia River estuary, showing inputs of river and coastal ocean water and particles and the location of the ETM at the head of the salt wedge. Curved arrows indicate mixing of freshwater (white) and seawater (dark gray). (B) Compositions of clone libraries at the phylum and subphylum levels. Arrows show movement of bacterial types from source populations into the estuary. Estuarine clone libraries are separated into clones unique to the estuary (bottom section of free-living and particle-attached charts), clones similar to those found in the river (upper left sections), and clones similar to those found in the coastal ocean (upper right sections). Estuarine clones were designated river or coastal ocean when they clustered with clones from these source communities. Most had at least 96% sequence similarity to river and coastal ocean clones.
Coastal ocean diversity.
Thirty-one of forty-five clones from the two coastal ocean libraries were closely related to coastal and marine clone sequences SAR11 and SAR7 (Sargasso Sea) (Fig. 2A and H), OM42 and -81 (North Carolina coast) (Fig. 2A and H), OCS124 and -155 (Oregon coast) (Fig. 2A and G), BAL47 (Baltic Sea) (Fig. 2B), and MED25 (Mediterranean Sea) (Fig. 2F); to marine isolates of Prochlorococcus spp. (Fig. 2H); and to NKB4 (deep sea sediment) (Fig. 2C). Six clones, potentially of terrestrial origin, were related to the soil isolate Planctomyces limnophilus, to the plant pathogen Pseudomonas syringae, and to Legionella spp. (Fig. 2E and C).
Clones related to the marine genera Prochlorococcus and Synechococcus were very common in the coastal ocean clone library (Fig. 2H). One cluster was very closely related to SAR7 (98.9 to 99.7%), an open-ocean clone related to Synechococcus spp. The other cluster was most closely related to low-light-adapted strains of Prochlorococcus spp. (97.3 to 98.6%) (MIT9303 and MIT9313) and less closely related to high-light-adapted strains (95.1 to 96.4%) (MIT9302 and MIT9312) (27).
The two coastal ocean clone libraries were very different (Table 3). The unfiltered coastal ocean clone library was dominated by cyanobacteria (35%), chloroplasts (22%), and γ-proteobacteria (22%). The clone library made with 3-μm-filtered water was dominated by α-proteobacteria (52%) and contained only three clones that were related to clones from the unfiltered clone library.
Estuarine diversity.
The free-living estuarine clone library was dominated by β-proteobacteria, gram-positive bacteria, α-proteobacteria, Cytophaga spp., and one type of γ-proteobacterium (Table 3). Twenty-one β-proteobacteria, one Verrucomicrobium clone, and seven gram-positive bacterial clones were related to clones from the river and belonged to clades of cosmopolitan freshwater bacteria or common soil bacteria (Fig. 2B, E, and G). Also, three α-proteobacterium clones and two gram-positive clones were related to clones found in the coastal ocean clone library. A total of 48% of free-living estuarine clones were related to clones isolated from the river or the coastal ocean (Fig. 3B).
Of the remaining 52% of free-living estuarine clones, all γ-proteobacterium clones, five Cytophaga clones, two Verrucomicrobium clones, and one δ-proteobacterium clone (30%) had no relatives in the river or the ocean clone library but were related to sequences in the particle-attached estuarine clone library (Fig. 2C to F). The remaining clones were unique to the free-living estuarine clone library.
The particle-attached estuarine clone library was dominated (75%) by clones that were rare in or absent from the river or the coastal ocean, including many clones related to Cytophaga spp. and α-proteobacteria and a diverse assemblage of γ-proteobacteria. Other particle-attached estuarine clones were related to clones in the particle-attached river library and the unfiltered coastal ocean library (Fig. 3B).
DISCUSSION
Bacterial diversity in the Columbia River estuary appears to be influenced by the rapid movement of water through the system and the trapping of particles in ETM. Water masses entering the estuary from the river and the coastal ocean are mixed by tidal action and are then washed out of the estuary at the surface, above the incoming layer of coastal marine water, in an average of 1 to 2 days (30) (Fig. 3A). The free-living bacterial communities associated with these water masses are also combined by tidal action and presumably wash out of the estuary just as rapidly. Clones isolated from the river and the coastal ocean generally fell into distinct freshwater or marine phylogenetic clusters (Fig. 2) and were similar to organisms and environmental clones isolated from other freshwater and marine systems. Nearly half of the free-living clones from the estuary were related to these freshwater and marine clones (Fig. 3B), demonstrating how this system acts as a mixing zone for bacterial communities and suggesting that free-living bacteria wash into and out of the estuary too rapidly to develop into an estuarine community.
The movement of particles and particle-attached bacteria in the estuary is very different from the movement of water. Allochthonous particles can be trapped in ETM by attaching to other particles, forming large, rapidly settling macroaggregates (35). In the ETM, these particles settle to the bed at slack tide and are resuspended during flood and ebb tides. The formation of these particles and their cycling in the ETM brings together both allochthonous and estuarine particle-attached bacteria. The particle-attached estuarine clone library showed evidence of this mixing in that it contained river and coastal ocean clones as well as uniquely estuarine clones (Fig. 3B).
We hypothesized that ETM promote the development of an estuarine bacterial community by trapping particles in the estuary. Particles trapped in ETM are thought to remain there for 2 to 4 weeks (35), creating within this fast-moving system a relatively stable estuarine environment in which estuarine organisms may have time to develop into a robust community. Estuarine clones unrelated to clones found in the river or the coastal ocean comprised 75% of the particle-attached clone library (Fig. 3B), suggesting that the particle-attached fraction of bacteria in the ETM was composed of organisms that developed in the estuary.
Particle-attached bacteria play a critical role in the ecosystem of the Columbia River estuary due to their relatively high activity and their high concentration in ETM. They are the most important decomposers of organic matter in the system, turning over particulate organic matter in an average of 8 to 71 days depending on the conversion efficiency (6). They are also important in the estuarine food web since they are directly consumed by detritivorous copepods (38) as well as rotifers and protozoa (5). We cannot say whether the allochthonous particle-attached organisms remain active in the estuary. However, if organisms unique to the particle-attached estuarine clone library did not wash in from other sources, then they must have actively developed in the estuary.
Clones unique to the estuary were found in both the particle-attached and free-living fractions, making it unclear whether these organisms were originally free-living or particle attached. However, the particle-attached fraction of bacteria had a much higher thymidine incorporation rate (Table 1) and a much higher level of extracellular enzyme activity (6), suggesting that uniquely estuarine organisms grew primarily on particles and were released into the free-living fraction in situ or perhaps during sample manipulation.
The largest groups of uniquely estuarine clones in the particle-attached fraction were related to Cytophaga spp., α-proteobacteria, and γ-proteobacteria. Environmental clones similar to Cytophaga spp., and γ-proteobacteria were also among the most abundant phylogenetic types found on marine snow particles in the Santa Barbara Channel (7). All but four of the Cytophaga-Flexibacter clones were found in the estuary, and most were in the particle-attached fraction (Fig. 2F). Cytophaga spp. exhibit gliding motility and are therefore thought to live primarily on surfaces. They are also known for their ability to produce exopolysaccharide slime and extracellular enzymes capable of degrading many different refractory biomacromolecules, including cellulose and chitin (36). Cytophaga spp. seem to be the ideal organisms to thrive as particle-attached bacteria in the estuary and may be among the hallmark bacterial types in the Columbia River estuary.
The largest cluster of clones in both the particle-attached and free-living estuarine clone libraries (17%) was most closely related to Marinomonas vaga (89.2 to 89.8%) and other members of the Oceanospirillum assemblage (Fig. 2C). There are many oceanic environmental clone sequences from other studies that appear to be related to this assemblage; however, most of these are only partial sequences that do not overlap with our sequences (NH16-1 and -18, NH29-6 and -17, NH49-13, and BDA1-8 and -10). Most genera in the Oceanospirillum and Alteromonas assemblages require NaCl for growth (23), but a subset can grow at the reduced salt concentrations typical of estuaries (11, 13). The sheer abundance of these clones in both estuarine libraries and the complete absence of them in the river and coastal ocean libraries suggests that they are estuarine organisms.
The study of environmental clone libraries is starting to reveal the existence of environment-specific clades of microorganisms, such as the recently described clades of cosmopolitan freshwater bacteria. This suggests that 16S rRNA diversity may reflect metabolic diversity. Two clusters of α-proteobacterium clones from this study provide examples of these environment-specific clades. Eleven clones were related to Rhodobacter spp. (Fig. 2A), a group that includes two clusters of phylogenetically distinct organisms from marine and freshwater environments (18). Members of the marine Rhodobacter group were recently shown to dominate coastal bacterioplankton communities, accounting for 28% of the rRNA genes in coastal ocean water collected off Sapelo Island, Georgia (14, 34). Members of the freshwater group have not been previously identified in environmental clone libraries but are known from culture.
Three groups of clones belonging to the SAR11 cluster (9, 12, 28) were identified, two marine and one freshwater. One clone collected in the Columbia River (CR-FL10) had a 100% sequence identity to LD12, a clone sequence collected from Lake Loosdrecht, The Netherlands. This clone is also closely related to lake clones ACK-M20 and ARC22 and others that comprise the freshwater SAR11 cluster. Marine clones from this cluster were very abundant in the coastal ocean clone library and were also found in the estuary.
Phenotypic capabilities cannot be determined directly from 16S sequences, but information about the environment and about related organisms in cultivation provides clues to the potential phenotypes of environmental clones. Some clusters of particle-attached clones from the Columbia River estuary were closely related to cultivated organisms with characteristics conducive to life on particles in ETM. Cultivated Rhodobacter spp. can grow aerobically and anaerobically, and many display some degree of halotolerence or osmotolerence (1, 16, 20, 21). δ-Proteobacteria include obligately anaerobic sulfate reducers and may grow in low-oxygen regions of ETM particles. Cytophaga spp., as described earlier, are surface-associated bacteria known to produce exopolysaccharides as part of biofilm formation and to release extracellular enzymes for the degradation of particulate organic matter. It is reasonable to hypothesize that clones related to these cultivated organisms share some of the same phenotypic capabilities. Bacteria with these known phenotypes probably comprise the most active fraction of bacteria on particles and may be an important component of the estuarine food web.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation land margin ecosystem research grant OCE-9412028.
We thank Charles A. Simenstad for tireless efforts as chief scientist on the Columbia River ETM project and the captain and crew of the RV Robert Gordon Sproul for assistance in the field.
REFERENCES
- 1.Abee T, Palmen R, Hellingwerf K J, Konings W N. Osmoregulation in Rhodobacter sphaeroides. J Bacteriol. 1990;172:149–154. doi: 10.1128/jb.172.1.149-154.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahr M, Hobbie J E, Sogin M L. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- 3.Baross J A, Crump B, Simenstad C A. Elevated ‘microbial loop’ activities in the Columbia River estuarine turbidity maximum. In: Dyer K R, Orth B J, editors. Changes in fluxes in estuaries: implications from science to management (ECSA22/ERF symposium, Plymouth, September 1992). Fredensborg, Denmark: Olsen & Olsen; 1994. pp. 495–464. [Google Scholar]
- 4.Berner E K, Berner R A. Global environment: water, air, and geochemical cycles. Englewood Cliffs, N.J: Prentice Hall; 1996. Marginal marine environments: estuaries; pp. 284–311. [Google Scholar]
- 5.Crump B C, Baross J A. Particle-attached bacteria and heterotrophic plankton in the Columbia River estuary. Mar Ecol Prog Ser. 1996;138:265–273. [Google Scholar]
- 6.Crump B C, Simenstad C A, Baross J A. Particle-attached bacteria dominate the Columbia River estuary. Aquat Microb Ecol. 1998;14:7–18. [Google Scholar]
- 7.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 8.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauthier M J, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, Bertrand J-C. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new extremely halotolerent, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol. 1992;42:568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- 11a.Gilbert, D. G. 11 July 1994, posting date. [Online.] SeqApp program, version 1.9a169. http://ftp.bio.indiana.edu/IUBio-software+Data/molbio/seqapp [27 May 1999, last day accessed.]
- 12.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 13.González J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol. 1997;47:369–376. doi: 10.1099/00207713-47-2-369. [DOI] [PubMed] [Google Scholar]
- 14.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen M C, Tolker-Nielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 16.Hansen T A, Imhoff J F. Rhodobacter veldkampii, a new species of phototrophic purple nonsulfur bacteria. Int J Syst Bacteriol. 1985;35:115–116. [Google Scholar]
- 17.Hiorns W D, Methé B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack Mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraishi A, Ueda Y. Intrageneric structure of the genus Rhodobacter: transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int J Syst Bacteriol. 1994;44:15–23. [Google Scholar]
- 19.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igeño M I, González del Moral C, Castillo F, Caballero F J. Halotolerence of the phototrophic bacterium Rhodobacter capsulatus E1F1 is dependent on the nitrogen source. Appl Environ Microbiol. 1995;61:2970–2975. doi: 10.1128/aem.61.8.2970-2975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imhoff J F. Halophilic phototrophic bacteria. In: Rodriguez-Valera F, editor. Halophilic bacteria. Vol. 1. Boca Raton, Fla: CRC Press; 1988. pp. 85–108. [Google Scholar]
- 22.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieg N R. Genus Oceanospirillum Hylemon, Wells, Krieg and Jannasch 1973, 361 (AL) In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 104–110. [Google Scholar]
- 24.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Methé B A, Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:386–374. [Google Scholar]
- 27.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 28.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 29.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.National Center for Biotechnology Information. 13 April 1997, revision date. [Online.] Programs. National Center for Biotechnology Information, Bethesda, Md. http://www.ncbi.nlm.nih.gov. [27 May 1999, last date accessed.]
- 30.Neal V T. Physical aspects of the Columbia River and its estuary. In: Pruter A T, Alverson D L, editors. The Columbia River estuary and adjacent ocean waters. Seattle: University of Washington Press; 1972. [Google Scholar]
- 31.Nold S C, Zwart G. Patterns and governing forces in aquatic microbial communities. Aquat Ecol. 1998;32:17–35. [Google Scholar]
- 32.Pedersen K, Arlinger J, Hallbeck L, Pettersson C. Diversity and distribution of subterranean bacteria in groundwater at Oklo in Gabon, as determined by 16S rRNA gene sequencing. Mol Ecol. 1996;5:427–436. doi: 10.1111/j.1365-294x.1996.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 33.Prahl F G, Small L F, Eversmeyer B. Biogeochemical characterization of suspended particulate mater in the Columbia River estuary. Mar Ecol Prog Ser. 1997;160:173–184. [Google Scholar]
- 34.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 35.Reed, D. J. Personal communication.
- 36.Reichenbach H. Genus 1. CytophagaWinogradsky 1929, 577, (AL) emend. In: Staley J T, Bryant M P, Pfenning N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams & Wilkins; 1989. pp. 2015–2050. [Google Scholar]
- 37.Simenstad C A, Small L F, McIntire C D, Jay D A, Sherwood C. Columbia River estuary studies: an introduction to the estuary, a brief history, and prior studies. Prog Oceanog. 1990;25:1–13. [Google Scholar]
- 38.Simenstad C A, Morgan C A, Cordell J R, Baross J A. Flux, passive retention, and active residence of zooplankton in Columbia River estuarine turbidity maxima. In: Dyer K R, Orth B J, editors. Changes in fluxes in estuaries: implications from science to management (ECSA22/ERF symposium, Plymouth, September 1992). Fredensborg, Denmark: Olsen & Olsen; 1994. pp. 473–484. [Google Scholar]
- 39.Small L F, McIntire C D, MacDonald K B, Lara-Lara J R, Frey B R, Amspoker M C, Wenfield T. Primary production, plant and detrital biomass, and particle transport in the Columbia River estuary. Prog Oceanogr. 1990;25:175–210. [Google Scholar]
- 40.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Wintzingerode F, Gobel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 45.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwart G, Hiorns W D, Methe B A, van Agterveld M P, Huismans R, Nold S C, Zehr J P, Laanbroek H J. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]
- 48.Zwart G, Huismans R, van Agterveld M P, Van de Peer Y, De Rijk P, Eenhoorn H, Muyzer G, van Hannen E J, Gons H J, Laanbroek H J. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol Ecol. 1998;25:159–169. [Google Scholar]