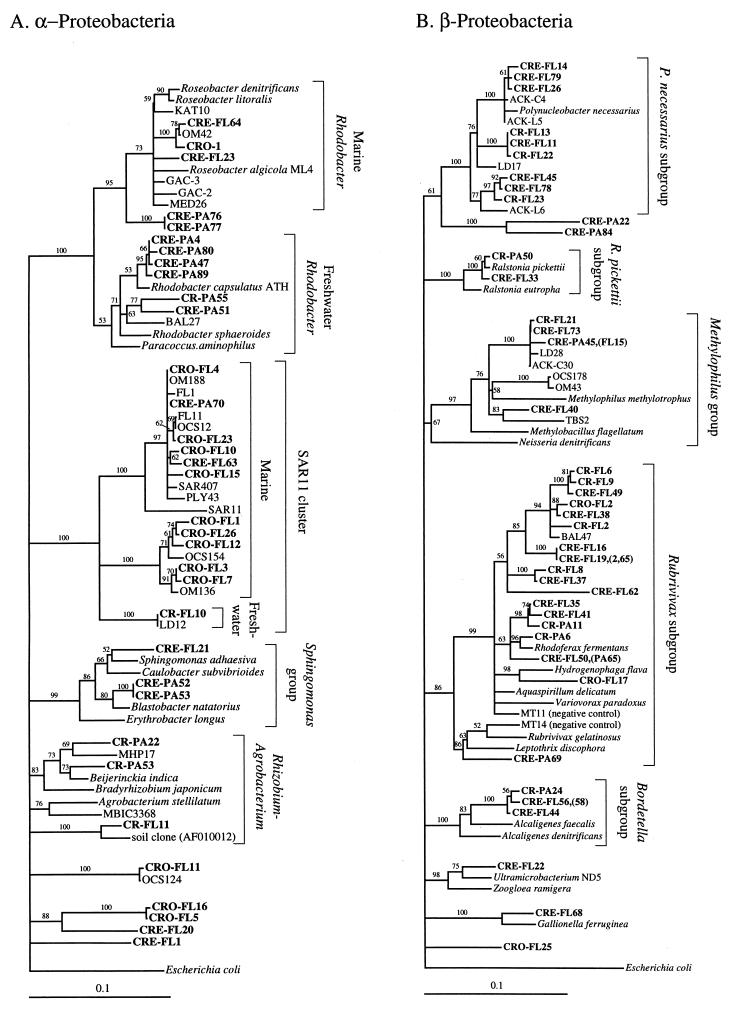
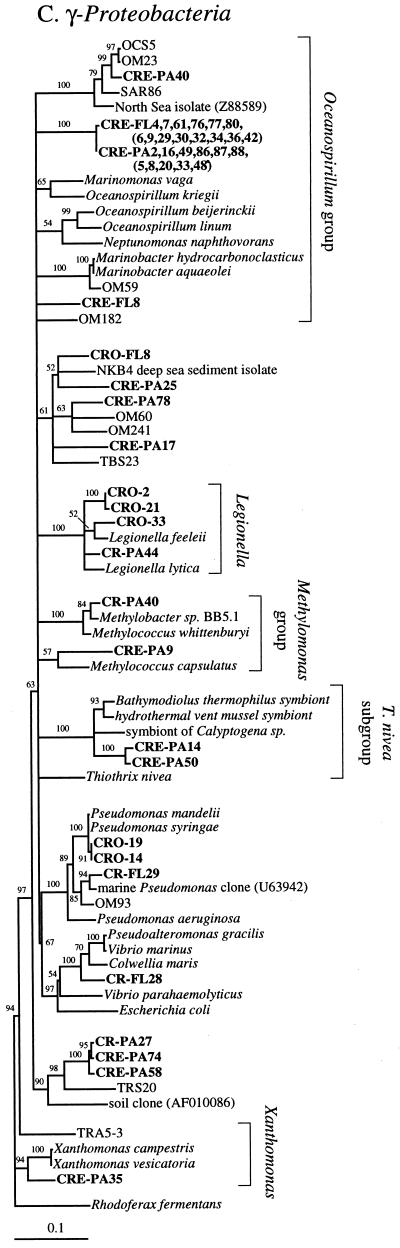
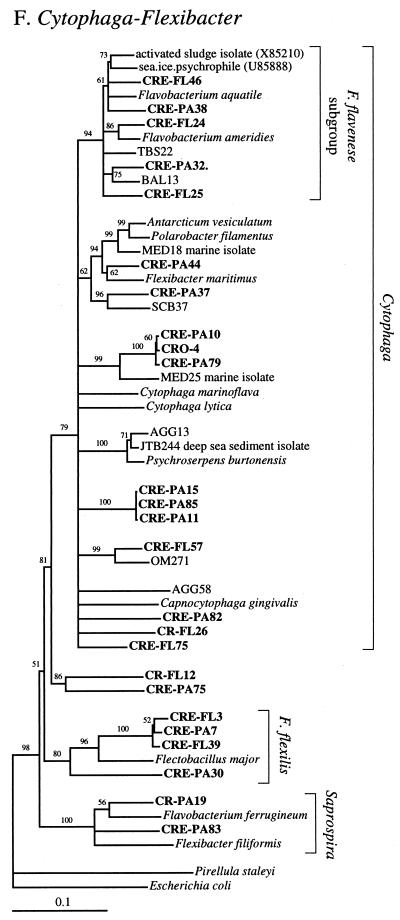
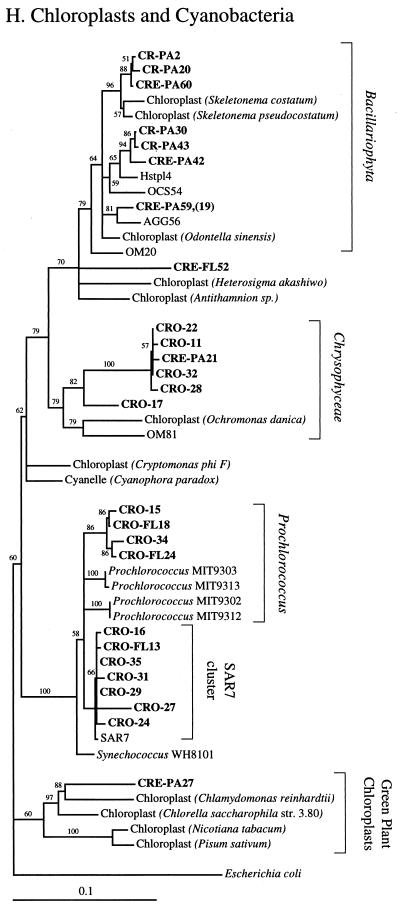
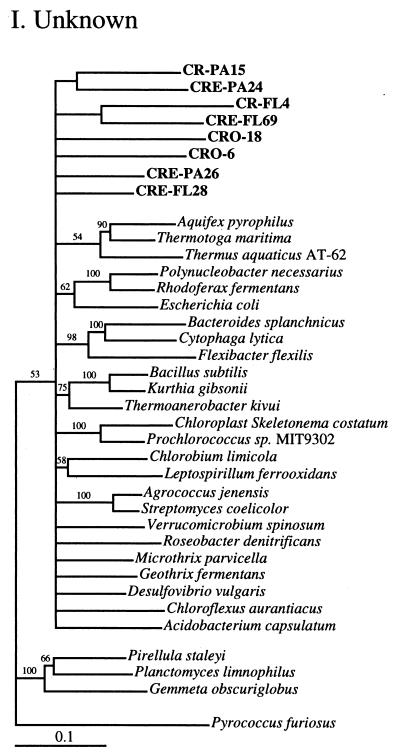
FIG. 2.
Phylogenetic relationships among 16S rRNA sequences from Columbia River, estuary, and adjacent coastal ocean clones and from other environmental clones and cultured organisms. (A) α-proteobacteria; (B) β-proteobacteria; (C) γ-proteobacteria; (D) δ-proteobacteria; (E) Verrucomicrobiales and Planctomyces clade; (F) Cytophaga-Flexibacter assemblage; (G) gram-positive bacteria; (H) chloroplasts and cyanobacteria; (I) all other clones. Fifty percent majority-rule trees were constructed by the neighbor-joining method. The percentages of 1,000 bootstrap replicates that supported the branching order are shown above or near the relevant nodes. The scale bars correspond to a 10% difference in nucleotide sequence. Clones from this study are indicated in boldface and are named with the following prefixes, designating their sources: CR, Columbia River; CRE, Columbia River estuary; CRO, coastal ocean; PA, particle attached; and FL, free-living. All sequences are available from the GenBank database, and accession numbers are provided if the organism or clone name is not unique. CFB, Cytophaga-Flexibacter-Bacteroides phylum.