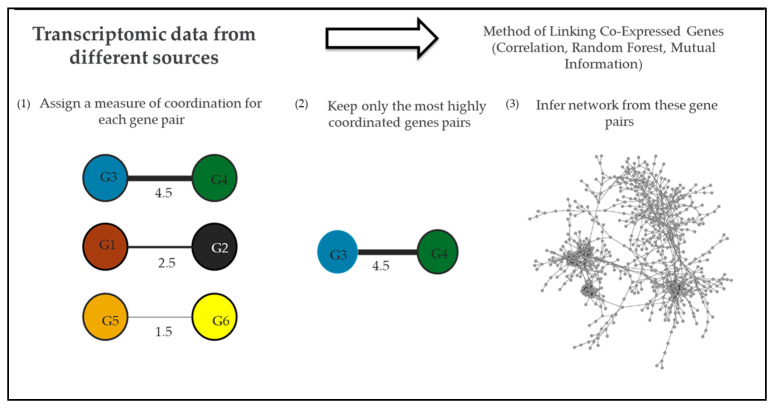
Figure 2.
Network analysis. To infer a gene co-expression network of a biological system transcriptomic data is first collected. Next, a network inference tool using correlation coefficient (e.g., Pearson), mutual information (e.g., Context Likelihood of Relatedness), random forest (e.g., GENIE3), or another method is used to calculate a co-expression value for each gene pair. Following this, only gene pairs that are highly co-expressed (either positively or negatively) are retained. Once this co-expression and filtering are done to all gene pairs a network of the most highly co-expressed genes can be inferred [96,100,101].