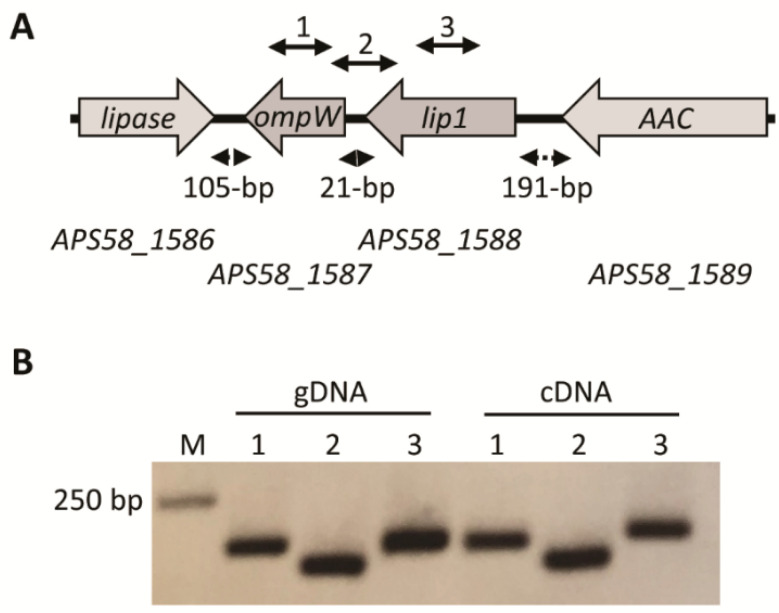
Figure 1.
The lip1-ompW operon. (A) Schematic organization of the operon and neighboring genes according to the annotation of the Acidovorax citrulli M6 genome (GenBank accession CP029373.1). Grey arrows represent the open reading frames (ORFs) and transcription direction of the following genes (numbers between parentheses indicate the position of start and stop codons in the A. citrulli M6 chromosome): APS58_1586, hypothetical protein but having high similarity to GDSL lipases (1,779,911–1,780,861); APS58_1587, outer membrane protein OmpW (1,780,966–1,781,658, complement); APS58_1588, Lip1 (1,781,679–1,782,830; complement); and APS58_1589, D-alanyl-D-alanine carboxypeptidase (AAC; 1,783,021–1,784,559; complement). Dashed double arrows indicate the distance between the ORFs. Solid double arrows above the genes indicate the location of PCR targets for cDNA amplification (see Table S1). (B) Polymerase chain reaction (PCR) using A. citrulli M6 genomic DNA (gDNA), or cDNA synthesized from RNA extraction from an overnight NB-grown culture of A. citrulli M6. PCR reactions were conducted with primers described in Table S1 and the lane numbers correspond to the solid double arrows in panel A: (1) primers ompW_F and ompW_R for amplification of an internal fragment of ompW; (2) primers 21between_F and 21between_R for amplification of a region containing part of the lip1 and ompW ORFs and their intergenic region; and (3) lip1_F and lip1_R for amplification of an internal fragment of ompW. Negative controls with no reverse transcriptase were used to verify that RNA samples do not contain genomic DNA contamination, and did not yield PCR products.