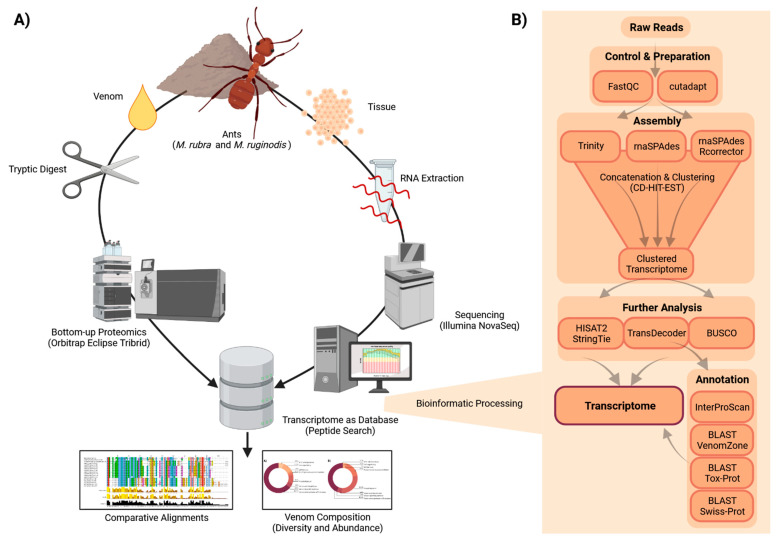
Figure 1.
Overview of our proteotranscriptomics workflow. (A) Proteomics workflow: Crude venom was collected, digested and analyzed in a bottom–up proteomics approach using an Orbitrap Eclipse Tribrid MS and the transcriptome database. (B) Transcriptomics workflow: RNA was sequenced on an Illumina NovaSeq system, and the raw sequencing data were preprocessed and assembled using multiple algorithms. The concatenated dataset was further analyzed and annotated based on different sources of information. The resulting ORFs were used as a database for the proteomics experiment. Transcripts validated at the proteome level were used for the subsequent analysis of venom components in both species.