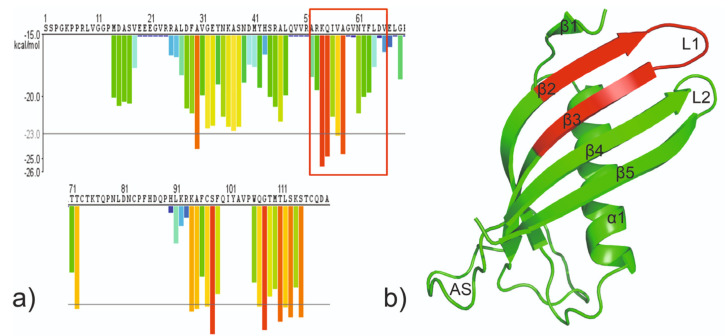
Figure 1.
(a) The 3D profile prediction of fibrillizing segments in HCC sequence. The predicted lowest energy template interactions according to ROSETTADESIGN energy calculation are plotted at the initial residue of each hexapeptide. Red and orange bars represent sequences of the hexapeptides which have tendency to form fibrils. Yellow, green, and blue bars represent sequences of the hexapeptides that are predicted not to form fibrils [25]. (b) The crystal structure of the monomeric mutant V57N (PDB: 3XN0; [18]) with marked fragments fulfilling potential conditions of steric zipper; red: Ala52 −Asp65.