Abstract
The protein HSF-1 is the controlling transcription factor of the heat-shock response (HSR). Its binding to the heat-shock elements (HSEs) induces the strong upregulation of conserved heat-shock proteins, including Hsp70s, Hsp40s and small HSPs. Next to these commonly known HSPs, more than 4000 other HSEs are found in the promoter regions of C. elegans genes. In microarray experiments, few of the HSE-containing genes are specifically upregulated during the heat-shock response. Most of the 4000 HSE-containing genes instead are unaffected by elevated temperatures and coexpress with genes unrelated to the HSR. This is also the case for several genes related to the HSP chaperone system, like dnj-12, dnj-13, and hsp-1. Interestingly, several promoters of the dedicated HSR-genes, like F44E5.4p, hsp-16.48p or hsp-16.2p, contain extended HSEs in their promoter region, composed of four or five HSE-elements instead of the common trimeric HSEs. We here aim at understanding how HSF-1 interacts with the different promoter regions. To this end we purify the nematode HSF-1 DBD and investigate the interaction with DNA sequences containing these regions. EMSA assays suggest that the HSF-1 DBD interacts with most of these HSE-containing dsDNAs, but with different characteristics. We employ sedimentation analytical ultracentrifugation (SV-AUC) to determine stoichiometry, affinity, and cooperativity of HSF-1 DBD binding to these HSEs. Interestingly, most HSEs show cooperative binding of the HSF-1 DBD with up to five DBDs being bound. In most cases binding to the HSEs of inducible promoters is stronger, even though the consensus scores are not always higher. The observed high affinity of HSF-1 DBD to the non-inducible HSEs of dnj-12, suggests that constitutive expression may be supported from some promoter regions, a fact that is evident for this transcription factor, that is essential also under non-stress conditions.
Subject terms: DNA, Transcription factors, Microarray analysis, Analytical biochemistry
Introduction
The heat shock transcription factor (Hsf) is the essential transcriptional activator of the heat-shock response (HSR). It activates the genes of the classical HSR like Hsp70, small HSPs and Hsp40s1–3. Hsf proteins are further involved in many developmental processes, like embryonic placenta development4, female meiotic division5, and general transcription6. Hsf proteins are also reported as negative regulators of RNA polymerase II promoters and modulate protein homeostasis, cellular proliferation7,8 and the regulation of multicellular organism growth9,10. While these functions are governed by the binding of Hsf proteins to HSEs distributed throughout the genome, they also dependent on the chromatin accessibility11, as well as the general environment in which these Hsf proteins are activated12,13. In the mammalian genome several Hsf-like proteins are encoded that control individual sets of target genes. In nematodes, the expressed, full-length Hsf-like gene (HSF-1) is encoded, while another Hsf-homolog gene, termed hsf-2, is represented by the pseudogene Y53C10A.3. With HSF-1 likely being the sole expressed and functional homolog of the Hsf-proteins, the interaction between HSF-1 and the many detected HSEs can be investigated without the necessity to differentiate between several Hsf-proteins14,15.
Despite this simplicity, the regulation of the heat-shock response in nematodes is complex, being influenced by the age of the animal and active mostly in muscular and intestinal tissues16. Further, larvae up to the L2 stage show a reduced expression of the HSR in contrast to older larval forms. The aging adult then is characterized by lower inducibility of the HSR17. These differences imply a complex regulation of HSF-1 activity during aging, which is thought to ensure that the nematode’s reproductive phase is best protected from stressful events18. Beyond that, nematode HSF-1 is participating in the innate immune response by upregulating specific target genes and in aging, where HSF-1 cooperates with the transcription factor DAF-1615,19–22. The observation that several thousand of HSEs are present in the promoter regions throughout the nematode genome, even though the canonical HSR seems to be restricted to few genes23,24, is puzzling. Several of the canonical heat-shock proteins, like HSP-90 are not even heat-inducible in C. elegans and also the canonical Hsp40-like proteins are not upregulated strongly upon heat-shock20,25.
HSF-1 in nematodes is a protein of 671 amino acids. Like other Hsf proteins, HSF-1 consist of several conserved domains, including the N-terminal DNA-binding domain (DBD), an oligomerization domain and a carboxyl-terminal regulatory domain. Nematode HSF-1 further contains an 82-amino-acids extension of unknown function at its N-terminus. Under normal growth conditions Hsf proteins are monomeric and form cytosolic complexes with Hsp90 and Hsc70. This interaction prevents the trimerization and activation of Hsf proteins. Under heat-stress or other inducing conditions, Hsf proteins are released from the protecting chaperones and oligomerize. In most cases Hsf binds as a trimeric protein to the HSE-containing DNA sequences. The phosphorylation of Hsf proteins triggers the translocation to the nucleus and initiates the transcription6,26–28. Despite these regulatory events, the interaction of Hsf proteins with consensus dsDNA is observable also for the non-activated Hsf. In this respect, it is mostly unclear, how Hsf proteins distinguish the various HSE-containing target genes.
Here we focus on the DNA binding domain of HSF-1 from C. elegans and aim at resolving its interaction with differently regulated HSEs from the nematode genome. To this end we first define the HSE-containing genes that are strongly upregulated upon heat-shock. We then use HSE-containing dsDNA constructs from these HSE regions to investigate to what extent the interaction parameters of the HSF-1 DBD with dsDNA are influenced by HSEs of different sequences and structural organization.
Material and methods
Analysis of microarray data
Initially three microarray data sets investigating the heat-shocked (GSM62937, GSM62941, GSM62945) versus non-shocked condition (GSM62936, GSM62940, GSM62944), which can be obtained from the GEO microarray depository under the GSE2862 tag31, were used to identify genes with strong overexpression. Here, L2 stage C. elegans larvae were heat-shocked for 20 min at 33 °C, followed by a recovery at 20 °C for 40 min. As expected, and previously published15, the strongest upregulated genes were hsp-16.1, hsp-16.48 and F44E5.4 and their duplicated loci. Individual genes that show elevated expression under heat-shock conditions were determined. To obtain information on whether these genes commonly express together, genome-wide clique set analysis was performed as described before29,30. The heat-shock data sets were used together with the publicly available coexpression cliques. Altogether 307 cliques had been obtained before, with the largest clique containing 1200 genes and the smallest clique containing 6 genes and the publicly available information was used (www.richterlab.de/DataSets/ and https://github.com/klarichter/clusterEX_cliques_Celegans)29,30. We then used each of the three microarray replicates to assign their values to the genes in the coexpression cliques and analyzed those in respect to significant induction or repression as previously described for yeast and nematode expression studies29,30. As these heat-shock data and the clique set were both based on the GPL200 platform (Affymetrix C. elegans genome st-1.0), each Probe Set ID was represented by exactly one value in the described clique set. Analyses were performed for each replicate and average values for each clique were calculated to rank the cliques according to their average induction and p-values for induction significance as described29,30.
Given the complexity of the heat-shock response, we compared these data to other genome-wide expression data sets. As such a heat-shock time course defined by microarray data32 was investigated as well as heat-shock experiments based on RNA sequencing15,17 . The Subio64 software package 1.24.5853 (Subio Inc. Kagoshima, Japan) was used to derive annotated, normalized expression data from the publicly available SRA-files in cases where the annotated data were not available from GEO repository.
HSE-detection in the promoter regions
HSE-detection in the promoter region 1000 bp upstream of the ATG was performed based on the PWM-models published for the human Hsf1’s DNA binding sequence33, which is represented by the following PWM pattern:
A | 0.41 -3.20 1.13 1.05 -0.86 -0.55 -2.10 -1.61 -0.17 -1.28 0.58 -5.77 1.14 1.11 0.26
C | -0.57 -5.19 -5.19 -3.58 0.84 0.73 0.02 0.34 1.14 0.06 -0.53 -5.19 -5.19 -5.19 -0.21
G | 0.84 1.70 -3.11 -1.20 -0.18 0.37 -0.70 -4.50 -0.27 -2.63 0.58 1.71 -5.19 -1.83 0.23
T | -5.77 -5.77 -5.77 -2.37 -0.08 -0.58 0.77 0.76 -2.40 0.79 -5.77 -5.77 -5.77 -5.77 -0.41
The 1000 bp promoter regions were obtained from Wormbase (www.wormbase.org)34 and searched with this HSF-1 consensus description. As recommended, a threshold level of 9 was used as lower limit for detection35. In several cases HSEs were detected in the same promoter and located only 5 bp from each other, which implies that the investigated HSE actually is a tetrameric HSE. If this pattern is observed a second time, a pentameric HSE-element was detected or in rare cases even larger arrays of HSEs were detected.
HSF-1 fragmentation and purification
Fragmentation was performed based on hydropathy plots and expression tests, which indicated that fragments, which contained additional domains outside the DBD showed either very weak expression or insoluble expression and that full-length HSF-1 could also not be obtained in soluble amounts sufficient for biochemical analysis. Due to our plan to investigate the direct interaction with the DNA, we chose the isolated DBD as model protein for interaction analysis. Therefore, the N- and C-terminus of C. elegans HSF-1 were determined by comparing both hydropathy plots and sequence alignments of different Hsf proteins from diverse species. This yielded the fragment AA82-AA198 which was subcloned into the pGATE vector (HSF-1 DBD) and thereby fused with a GST-tag. A GST-trap column was used for purification and the GST-tag was cleaved off by TEV-protease before the HSF-1 DBD was further purified via ion exchange chromatography and size-exclusion chromatography (all columns from GE Healthcare, Chicago, USA). Purity was determined by SDS-PAGE and peptide fingerprinting using mass spectrometry on a Bruker ultra-flex III MALDI-TOF/TOF instrument (Bruker, Billerica, USA) was employed to confirm the identity of the protein.
Circular dichroism spectroscopy
CD-spectroscopy on a Jasco J-715 was performed to obtain information on the structure and stability of the HSF-1 fragment. The folding state and the thermal stability of the expressed HSF-1 fragment was assessed at a concentration of 0.2 mg/mL in storage buffer (40 mM K2HPO4, 150 mM KCl). CD-spectra were recorded in the Far-UV region between 215 and 260 nm. To analyze the thermal stability of the fragment an unfolding transition was recorded at 220 nm in a temperature range between 25–95 °C.
Thermal shift assays
The stability of the folded structure was analyzed with thermal shift assays in a Mx3005P qPCR cycler (Stratagene, La Jolla, USA). Thermal shift assays were performed at a protein concentration of 0.2 mg/mL after addition of SYPRO orange (Invitrogen, Waltham, USA) at a dilution of 1:1000. The total volume was adjusted to 20 µL with storage buffer. The emission of SYPRO orange is recorded by excitation at 470 nm at a wavelength of 570 nm to monitor the temperature induced transition in the temperature range of 25 to 95 °C.
EMSA shift assays
ssDNA probes representing the promoter regions of F44E5.5, hsp-16.2a, hsp16.2b, hsp-1, hsp-70, dnj-12 and dnj-13 were obtained from Eurofins MWG Biotech (Eurofins MWG Biotech, Ebersberg, Germany). An equal amount of forward and reverse complementary strand was incubated at 95 °C and aligned at room temperature. The interaction between these dsDNAs and the HSF-1 DBD was then monitored by EMSA shift assays, by performing a native PAGE after DBD was added to the dsDNA. Gels were incubated in SYBR green for DNA detection and analyzed in a Typhoon Fluorescence Scanner (GE Healthcare, Chicago, USA) using the Alexa Fluor filter at 532 nm and stained with Coomassie for visualization of the protein complex.
Analytical ultracentrifugation and determination of species distributions
Analytical ultracentrifugation was performed in a ProteomeLab XL-A analytical ultracentrifuge (Beckman-Coutler, Brea, USA) to determine the binding of HSF-1 DBD to dsDNA sequences. To this end single strand DNA sequences from different promoter regions of the same length were mixed with equal amounts of their complement strand in storage buffer, heated up to 95 °C and then cooled to RT to generate the double stranded DNA product that represents the promoter region. HSF-1 DBD was added to 1.5 µM dsDNA at different concentrations (0 µM, 2.25 µM, 4.5 µM, 7.5 µM, 10.5 µM, 15 µM and 22.5 µM) and the absorbance of these samples was detected in analytical ultracentrifugation sedimentation velocity experiments at 260 nm and 280 nm at 42,000 rpm.
Data analysis of individual samples was performed with UltraScan III Version 4.0 (https://ultrascan3.aucsolutions.com/)36. All experiments were analyzed with the 2DSA-IT model employing the same settings (s-value range from 0 to 10 and f/f0 range from 1 to 4). This way two species distributions were obtained for each experiment, one for the data at 280 nm and one for 260 nm. The complexity of these distributions did not allow a unanimous assignment of solutes to species, which suggests that for a unifying solution a further reduction in search space has to be enforced. A reduced model therefore contained only the most abundant species of the binding reaction (HSF-1 DBD, ssDNA, dsDNA, dsDNA + 1 HSF-1, dsDNA + 2 HSF-1, dsDNA + 3 HSF-1, dsDNA + 4 HSF-1 and dsDNA + 5 HSF-1) at defined s20,w values. These values were known for HSF-1 DBD, dsDNA and ssDNA from control experiments, while the other species were estimated from a stepwise optimization of these values. Given that all DNA strands were of the same size, a unique value for the sedimentation coefficient (s20,w) of each assembly intermediate was assigned independent of the dsDNA used.
A custom grid model containing the species at the respective s20,w values was developed in UltraScan III and used to fit all data sets again. RMSD values of the unconstrained fit and the custom grid constrained fit were compared to verify that the fit quality despite the constraints is acceptable and the species s20,w values are sufficiently good estimates. To estimate the specific volume of each species and to confirm the MW of each obtained species the following equation was used:
Value pairs for D20,w and s20,w were estimated and the extinction coefficients, specific volumes and molecular weights were calculated for each species in the custom grid model.
Estimation of interaction parameters for dsDNA-DBD interaction
Data analysis was finally performed using the species concentrations determined from UltraScan III in the first unconstrained 2DSA-IT analysis and data fitting was based on previously developed models. The fitting function was modified from an Origin DLL-file developed originally for the interaction of two proteins (PPH-5 and HSP-90)37 to now describe the five-step binding process. Fitting was performed in analogy to the Nelder-Mead implementation for C# accessible at https://docs.microsoft.com/de-de/archive/msdn-magazine/2013/june/test-run-amoeba-method-optimization-using-csharp. Employing this function, KD-values for each step could be estimated. To this end, detected species absorbance was converted to species concentrations by employing the estimated extinction coefficients and fitting was performed globally for all species at both wavelengths. In few cases, especially where binding was very weak, RMSD values were almost exclusively influenced by the free ligand concentration. Under these conditions a weighing factor of 0.3 or 0.1 was applied to the free HSF-1 concentration to give more relevance to the other species M, ML1, ML2, ML3, ML4 and ML5. Cooperativity was observed, if KD-values for later assembly steps showed higher affinity than KD-values for early binding steps. Despite the constrained model the obtained KD-values contain large error intervals and are therefore considered as estimations due to the complexity of the binding events and the differences within the individual binding sites on the DNA.
ChIPseq-data analysis
ChIPseq data as available from the GEO repository were obtained as bedgraph-files17. Bedgraph files were searched to retrieve the values for specified regions and the reads identified in HSF-1 IP under various conditions were summarized in Excel to display the regions relevant for the genes of interest.
Results
The heat-shock response is represented by a small set of genes in C. elegans
We initially aimed at identifying those HSE-containing promoters that are most strongly upregulated under heat-stress conditions. Given that the HSR is complex in nematodes we used data from several heat-shock studies based on microarray and RNAseq analysis. Besides defining the individual genes, which are induced upon heat-shock in the different experiments, we also tested, whether the differently regulated genes are enriched in one or more of the 307 C. elegans coexpression cliques. These groups of genes (or gene sets) were obtained based on coexpression analysis of more than 2000 microarray experiments recently30 and found to contain many coexpressing tissue specific, phenotype specific and GO-term specific gene sets.
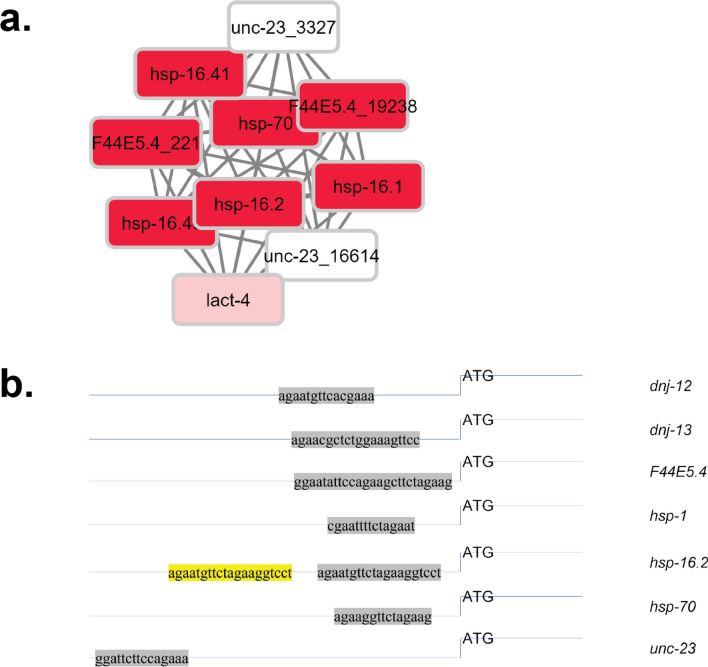
To initially define heat shock inducible and non-inducible HSE-sites in the promoter regions from the experiment series GSE286231, we defined the gene sets (or cliques) that represent the heat-shock response under these conditions (L2 larvae, 20 min 33 °C followed by recovery period of 40 min). To this end, we used the method previously described30 to determine significant coregulation units responding to heat shock. The procedure searches the 307 predefined coexpression cliques and identifies those with significant expression changes. In all three replicates of GSE286231 in particular one gene set out of the 307 cliques was highly induced (log2 > 2), the clique termed hsp-16.2-F44E5.4_19238, which contains the well described heat-shock genes hsp-16.1, hsp-16.2, hsp-16.48, hsp-16.41, hsp-70, F44E5.4 and F44E5.5 and in addition unc-23 and lact-4 (replicate 1 in Fig. 1a, Summary of the three replicates in Table 1, whole genome clustered in Supplemental Fig. 1). While unc-23 and lact-4 were not significantly upregulated in the three microarray experiments, the other genes of this coregulation clique are highly induced so that the hsp-16.2-F44E5.4_19238 gene set stands out with a 4.2-fold average induction (Table 2). Several canonical chaperones, like dnj-12 (two probes in cliques cdc-42_17192-rab-5_18073 and srj-42-srw-113), dnj-13 (two probes in cliques unc-116_2109-zfp-1_3976 and tars-1-AFFX-r2-3026-5_at) and the constitutively expressed Hsc70-homolog hsp-1 (clique dld-1-skn-1_16701) are not part of the HSR-clique hsp-16.2-F44E5.4_19238 and we individually tested their induction to confirm that they are indeed not coregulated with the induced heat-shock proteins (Table 2). As we find them them not upregulated in either of the replicates, the assignment to other coexpression cliques seems justified..
Figure 1.
Determined highly induced coexpression cluster and utilized promoter regions. (a) The highly induced clique of the genome clustered in 307 expression groups derived from public expression experiments. The color code reflects the heat-shock response as determined by Wang et al. (GSE2862)31. The highly induced cluster is enlarged for better visualization, while the whole clique map of the genome can be found in Supplemental Fig. 1. (b) Promoter region of inducible and non-inducible chaperones, relative to the ATG start codon.
Table 1.
Cliques identified as significantly up- or downregulated in the heat-shock experiments of Wang et al. (GSE2862)31.
| Clique number | Clique name | Clique size | Genes with signal in array | Average log2 | STD log2 |
|---|---|---|---|---|---|
| 268 | hsp-16.2-F44E5.4_19238 | 10 | 10 | 2.06 | 0.37 |
| 5 | Y73B3A.2_1097-Y73B3A.2_1130 | 17 | 2 | 0.33 | 0.71 |
| 104 | C10A4.6-C01A2.6 | 16 | 16 | 0.27 | 0.14 |
| 291 | fbxc-6-F09C6.1 | 8 | 8 | 0.24 | 0.36 |
| 163 | R05H5.4-F22D6.9_20949 | 5 | 5 | 0.22 | 0.27 |
| 235 | C10C5.3-C10C5.5 | 6 | 6 | 0.22 | 0.20 |
| 241 | fbxa-154_11499-Y44A6D.1 | 74 | 67 | 0.22 | 0.12 |
| 250 | K12H6.6_1575-C23H4.6 | 6 | 6 | 0.19 | 0.33 |
| 80 | C17H1.6-C17H1.13 | 76 | 75 | 0.19 | 0.13 |
| 169 | C33H5.1-F49C12.2 | 11 | 11 | 0.18 | 0.36 |
The average values for the three replicate experiments are shown together with the standard deviation and the clique size.
Table 2.
Significantly enriched genes and their clique assignment, or number of HSEs in the promoter region of the included genes.
| Gene name | Average expression change log2 | STD | Member of clique | Controlled by HSEs [number] |
|---|---|---|---|---|
| hsp-16.41 | 4.33964945 | 0.47257324 | hsp-16.2-F44E5.4_19238 | 7 |
| hsp-16.1 | 3.37025306 | 0.36880836 | hsp-16.2-F44E5.4_19238 | 7 |
| hsp-16.2 | 3.24598022 | 0.57114605 | hsp-16.2-F44E5.4_19238 | 7 |
| F44E5.4_221 | 2.43535939 | 0.6531605 | hsp-16.2-F44E5.4_19238 | 10 |
| hsp-16.48 | 2.43449799 | 0.26151971 | hsp-16.2-F44E5.4_19238 | 7 |
| F44E5.4_19238 | 2.42823859 | 0.70781857 | hsp-16.2-F44E5.4_19238 | 10 |
| C27D6.1 | 2.17492883 | 0.66776873 | 176662_at-Y53F4B.16 | – |
| fbxb-22 | 2.1705405 | 1.27504799 | sdz-10-fbxb-62 | – |
| math-6 | 2.12527449 | 0.85966164 | srj-21-srh-32 | – |
| F20B6.7 | 1.95293402 | 0.75883293 | tub-2_4713-daf-2_5288 | – |
| C15B12.3 | 1.87784369 | 0.59022648 | 172183_at-176110_at | – |
| Y6B3B.2 | 1.79628476 | 1.35642504 | 183712_at-srbc-74_8864 | – |
| F28A10.9 | 1.7697348 | 0.4485698 | srj-42-srw-113 | – |
| C01B4.2 | 1.74098252 | 0.42244849 | srj-21-srh-32 | – |
| str-243 | 1.72570157 | 0.42783197 | sre-33-ZK1025.1_8337 | – |
| hsp-70 | 1.69538497 | 0.43818876 | hsp-16.2-F44E5.4_19238 | 3 |
| C07A9.5_18941 | 1.68623723 | 1.47072224 | F28H1.5-T05D4.5 | – |
| Y16E11A.2 | 1.67500495 | 0.73573836 | fbxa-154_11499-Y44A6D.1 | 12 |
| C31G12.3 | 1.64731562 | 1.2334763 | srj-42-srw-113 | – |
| F07B7.8_11449 | 1.62495315 | 1.43400867 | 183712_at-srbc-74_8864 | – |
| clec-142 | 1.61733446 | 1.76090815 | F28H1.5-T05D4.5 | – |
| srh-258 | 1.59055877 | 0.53259038 | srj-42-srw-113 | – |
| scl-19 | 1.59004684 | 1.03454503 | srj-42-srw-113 | 3 |
| Y68A4B.3 | 1.58552192 | 1.18455696 | C33H5.1-F49C12.2 | – |
| clec-20 | 1.58507895 | 0.97920764 | sre-33-ZK1025.1_8337 | – |
| wrt-7 | 1.58082879 | 0.50613839 | grl-19-C13A2.4 | – |
| C04H5.7 | 1.56608678 | 0.85708337 | C46C2.5_15926-W03F11.1 | – |
| fbxb-118 | 1.56221989 | 0.81840697 | sdz-10-fbxb-62 | – |
| pqn-16 | 1.5514195 | 1.07311037 | srj-42-srw-113 | – |
| srh-154_13942 | 1.53974391 | 1.00629988 | srj-21-srh-32 | 2 |
| … | ||||
| lact-4 | 0.48976226 | 0.47729707 | hsp-16.2-F44E5.4_19238 | – |
| dnj-12_21104 | 0.2791495 | 0.14801702 | cdc-42_17192-rab-5_18073 | 3 |
| hsp-1 | 0.23010296 | 0.59319525 | dld-1-skn-1_16701 | 6 |
| unc-23_16614 | 0.0047227 | 0.216626 | hsp-16.2-F44E5.4_19238 | 6 |
| unc-23_3327 | − 0.07377314 | 0.30315485 | hsp-16.2-F44E5.4_19238 | 6 |
| dnj-13_17082 | − 0.41124351 | 0.31596264 | tars-1-AFFX-r2-3026-5_at | 7 |
| dnj-12_3674 | − 0.49049134 | 0.41234554 | srj-42-srw-113 | 3 |
| dnj-13_3319 | − 0.51080312 | 0.14089521 | unc-116_2109-zfp-1_3976 | 7 |
Individual induced genes are shown and their association to cliques. Further the number of HSEs modules that can be found within 1000 bp of their upstream promoter sequence.
Nematode HSEs vary widely in size and co-expression clique affiliation
We aimed at understanding, whether different affinities of the heat-shock transcription factor HSF-1 for the promoter sequences can be observed. Previous reports had highlighted that large number of HSEs can be found in the nematode genome24,38. Most of these genes are not induced in the heat-shock experiment investigated here. To obtain the HSEs of the genes of interest we searched the 1000 bp promoter regions of all genes of C. elegans. We identified 4120 HSE in genes, which contain a consensus sequence for HSF-1 in their promoter region. Despite not being induced upon heat-shock, several genes related to the chaperone system were found to contain HSE-like sequences in their promoter region, like dnj-12, dnj-13, and hsp-1. We then compared the sequence and structure of the HSEs in the promoter region of the chaperone proteins. Here, several promoters in the HSR-cluster contain more HSEs than the usually expected trimeric DNA-binding sequence, like hsp-16.2a and F44E5.4, which contain four or five HSF-1 binding sites in close vicinity (Fig. 1b).
Heat-shock inducibility varies with the employed stress conditions
We used data from other heat-shock experiments—performed with RNAseq—to see, whether these chaperones and heat-shock proteins are induced with the same pattern. In these RNAseq experiments analysis had been performed in young adults and L2 larvae with and without a heat-shock exposure. In the experiment performed by Brunquell et al.15, a very similar set of genes was induced upon heat-shock and likewise only one coexpression clique out of the 307 was found to be significantly upregulated, the clique hsp-16.2-F44E5.4_19238. Concomitantly the chaperone genes also represent the strongest upregulated genes on the single-gene basis (Table 3, Supplemental Fig. 2a). This also was observed in the second RNAseq experiment performed by Li et al.17 in L2 and young adult larvae (Table 4, Supplemental Fig. 2b).
Table 3.
Significantly enriched genes in the heat-shock experiments of Brunquell et al.15.
| GeneName | Mean_noHS | Mean_HS | LOG_Ratio |
|---|---|---|---|
| F44E5.5 | 9.747 | 1706.353 | 7.452 |
| F44E5.4 | 9.781 | 1703.300 | 7.444 |
| R11A5.3 | 0.478 | 63.026 | 7.044 |
| hsp-16.2 | 5.804 | 754.015 | 7.021 |
| hsp-16.41 | 8.159 | 944.460 | 6.855 |
| hsp-70 | 1.577 | 154.207 | 6.612 |
| nspe-2 | 0.459 | 42.666 | 6.539 |
| hsp-16.11 | 24.686 | 2280.832 | 6.530 |
| hsp-16.1 | 27.345 | 2520.684 | 6.526 |
| nspe-1 | 1.575 | 125.482 | 6.316 |
| MTCE.8 | 0.754 | 53.406 | 6.147 |
| hsp-16.49 | 29.527 | 1910.189 | 6.016 |
| hsp-16.48 | 28.702 | 1855.322 | 6.014 |
| K02F2.11 | 0.338 | 17.087 | 5.658 |
| F26D10.24 | 0.330 | 12.838 | 5.282 |
| MTCE.7 | 132.948 | 5111.687 | 5.265 |
| ostf-4 | 0.303 | 6.745 | 4.477 |
| 21ur-14804 | 3.447 | 65.291 | 4.244 |
| 21ur-6528 | 3.807 | 67.278 | 4.143 |
| col-84 | 0.346 | 4.882 | 3.819 |
| Y17D7C.2 | 0.296 | 3.799 | 3.682 |
| C13A2.12 | 0.341 | 4.320 | 3.663 |
| MTCE.17 | 0.383 | 4.713 | 3.622 |
| 21ur-14438 | 0.286 | 3.474 | 3.602 |
| MTCE.27 | 0.291 | 3.410 | 3.549 |
| rpr-1 | 0.000 | 17.693 | Inf |
| 21ur-8810 | 0.000 | 11.528 | Inf |
| lfor-1 | 0.000 | 8.067 | Inf |
| lfor-2 | 0.000 | 7.890 | Inf |
| 21ur-4739 | 0.000 | 10.792 | Inf |
| T06E8.3 | 0.000 | 3.935 | Inf |
| nspe-5 | 0.000 | 6.856 | Inf |
| …. | |||
| unc-23 | 18.625 | 75.578 | 2.021 |
| dnj-13 | 73.991 | 188.783 | 1.351 |
| hsp-1 | 606.752 | 1026.459 | 0.758 |
| dnj-12 | 106.633 | 84.553 | -0.335 |
Individual induced genes are shown compared to their non-stressed state. To define, which genes are expressed, it is to be considered that the median count is 7.7 and 5.2 for the two conditions investigated.
Table 4.
Significantly enriched genes in the heat-shock experiments of Li et al.17.
| GeneName | Mean_noHS | Mean_HS | LOG_Ratio |
|---|---|---|---|
| hsp-16.41 | 1.139 | 7114.820 | 12.609 |
| hsp-16.2 | 1.414 | 7361.540 | 12.346 |
| hsp-70 | 0.188 | 558.244 | 11.539 |
| R11A5.3 | 0.810 | 1052.930 | 10.344 |
| ZC21.10 | 1.134 | 378.419 | 8.382 |
| R107.5 | 30.439 | 3524.790 | 6.855 |
| C25F9.2 | 0.068 | 5.761 | 6.411 |
| dct-8 | 0.483 | 32.069 | 6.054 |
| nurf-1 | 63.938 | 4236.550 | 6.050 |
| fbxb-72 | 0.122 | 7.221 | 5.886 |
| Y71G12B.18 | 0.153 | 8.825 | 5.851 |
| scp-1 | 15.798 | 863.127 | 5.772 |
| F53A9.2 | 1.313 | 64.584 | 5.620 |
| tts-1 | 9.980 | 446.779 | 5.484 |
| Y43F8B.2 | 63.126 | 2546.830 | 5.334 |
| dpy-17 | 2.135 | 78.616 | 5.202 |
| Y38H6C.8 | 1.134 | 33.264 | 4.875 |
| col-106 | 0.049 | 1.414 | 4.852 |
| bath-34 | 0.441 | 12.651 | 4.843 |
| mtl-1 | 1.301 | 35.785 | 4.782 |
| nspc-12 | 0.568 | 15.630 | 4.782 |
| R05D7.2 | 0.049 | 1.308 | 4.741 |
| D2045.5 | 0.045 | 1.123 | 4.646 |
| ugt-14 | 0.275 | 6.485 | 4.561 |
| C30E1.9 | 139.027 | 3245.950 | 4.545 |
| C01G12.14 | 0 | 74.4564 | Inf |
| C09B8.12 | 0 | 14.777 | Inf |
| C11G10.11 | 0 | 16.9277 | Inf |
| C15H7.8 | 0 | 14.8235 | Inf |
| C16A3.15 | 0 | 19.6119 | Inf |
| C17H12.34 | 0 | 16.3819 | Inf |
| C25E10.14 | 0 | 1.71674 | Inf |
| …. | |||
| unc-23 | 52.588 | 326.964 | 2.636 |
| dnj-13 | 161.747 | 792.283 | 2.292 |
| hsp-1 | 1050.010 | 2194.130 | 1.063 |
| dnj-12 | 217.845* | 232.39* | 0.093 |
Individual induced genes are shown compared to their non-stressed state. *The gene dnj-12 was not included in the available final tables and therefore was calculated based on the raw data. The raw data values were divided by 40 to achieve a similar level of normalization as for the other genes. Raw data mean prior to normalization for all genes is 80 versus 83, thus substantially lower compared to the values of 8713.66 and 9295.77 found for dnj-12 prior to normalization.
We inspected one other experiment32, which had determined a time course of the heat-shock response, to investigate whether further genes get differentially expressed after prolonged incubation at the heat-shock temperature. At the shortest incubation time, hsp-16.2-F44E5.4_19238 was the dominant differentially expressed gene set and the chaperones in this clique were the genes with the strongest expression changes (Table 5, Supplemental Fig. 2c, Clique set in Supplemental Fig. 3a). This changes with longer exposure times and after 720 min of heat-shock several cliques are differentially expressed representing gene groups from very different processes and tissue specific expression (Table 5, Supplemental Fig. 2d, Clique set in Supplemental Fig. 3b). The cliques identified differ in their kinetics to heat-stress, in that most are not substantially affected at the shortest heat-exposure (30 min), but get affected starting from 60 min incubation time (Supplemental Fig. 4). Of the genes expressed under the harshest conditions, only few contain HSEs in their promoter region and even under those conditions dnj-12, dnj-13 and hsp-1 are only weakly changing their expression levels, while the heat-shock genes grouped in hsp-16.2-F44E5.4_19238 are highly elevated at all time points. Therefore, we consider these HSE-regulated genes to be “heat-inducible” while dnj-12, dnj-13 and hsp-1 represent genes that change their expression more weakly under heat-shock, despite HSE-sequences in the promoter region. unc-23, despite having been assigned to the HSR coexpression clique hsp-16.2-F44E5.4_19238 by the global coregulation analysis, also is upregulated weaker compared to the small heat-shock proteins and the Hsp70s.
Table 5.
Significantly enriched genes in the heat-shock experiments of Jovic et al.32.
| Gene name | 0 min level | 30 min-0 min | 60 min-0 min | 120 min-0 min | 180 min-0 min | 240 min-0 min | 360 min-0 min | 480 min-0 min | 720 min-0 min |
|---|---|---|---|---|---|---|---|---|---|
| hsp-16.41 | 9.36 | 15.27 | 15.45 | 15.12 | 16.79 | 17.21 | 16.50 | 16.20 | 16.11 |
| hsp-16.2 | 8.74 | 14.25 | 14.48 | 14.43 | 16.22 | 16.84 | 15.60 | 15.39 | 15.15 |
| F44E5.5 | 11.51 | 16.72 | 16.80 | 16.42 | 17.52 | 18.09 | 17.20 | 16.65 | 16.09 |
| T27E4.8 | 10.31 | 15.48 | 15.85 | 15.43 | 16.51 | 17.23 | 16.50 | 15.95 | 15.79 |
| T27E4.3 | 9.31 | 14.32 | 14.13 | 14.15 | 15.50 | 16.20 | 15.42 | 15.34 | 15.17 |
| hsp-16.41 | 8.71 | 13.15 | 13.59 | 13.48 | 15.26 | 15.79 | 14.43 | 14.68 | 14.71 |
| hsp-70 | 10.35 | 13.92 | 15.69 | 14.98 | 16.67 | 16.95 | 16.33 | 15.88 | 15.34 |
| F44E5.5 | 10.54 | 13.77 | 14.37 | 14.35 | 15.57 | 16.03 | 15.62 | 15.72 | 15.52 |
| skr-15 | 6.19 | 9.15 | 6.65 | 6.91 | 6.08 | 6.63 | 6.24 | 6.37 | 6.59 |
| C18A3.5 | 7.57 | 10.41 | 8.24 | 8.68 | 8.39 | 8.81 | 8.60 | 8.90 | 8.95 |
| T10D4.2 | 6.67 | 9.51 | 6.61 | 6.97 | 6.23 | 6.95 | 6.47 | 6.78 | 6.93 |
| M117.5 | 6.34 | 8.58 | 6.42 | 6.81 | 6.44 | 6.71 | 6.23 | 6.55 | 6.58 |
| nurf-1 | 12.27 | 14.36 | 16.72 | 15.82 | 17.33 | 17.13 | 16.90 | 16.70 | 15.92 |
| ZK616.3 | 9.33 | 11.33 | 9.21 | 9.77 | 9.64 | 9.82 | 9.34 | 9.58 | 9.80 |
| BJ140948 | 6.60 | 8.57 | 10.30 | 10.43 | 11.89 | 12.23 | 11.90 | 11.70 | 11.73 |
| nurf-1 | 11.41 | 13.32 | 16.12 | 14.92 | 16.35 | 16.56 | 16.32 | 16.17 | 15.24 |
| C08G5.7 | 6.00 | 7.83 | 6.14 | 6.30 | 5.85 | 6.22 | 5.82 | 6.08 | 5.89 |
| F19B2.5 | 11.62 | 13.42 | 14.92 | 14.34 | 15.32 | 15.48 | 14.65 | 14.68 | 14.53 |
| C25F9.6 | 7.81 | 9.54 | 10.97 | 10.42 | 11.38 | 11.59 | 11.10 | 11.52 | 10.97 |
| aip-1 | 10.29 | 12.01 | 13.83 | 13.13 | 14.19 | 14.23 | 13.48 | 13.23 | 12.92 |
| F59C12.4 | 11.51 | 13.22 | 13.43 | 13.69 | 15.84 | 15.26 | 15.69 | 15.99 | 15.87 |
| F59C12.4 | 11.84 | 13.52 | 13.89 | 14.04 | 16.10 | 15.62 | 16.03 | 16.37 | 16.14 |
| Y94H6A.10 | 11.96 | 13.63 | 14.01 | 14.20 | 15.21 | 15.11 | 14.86 | 15.03 | 14.94 |
| F59C12.4 | 11.99 | 13.61 | 14.07 | 14.20 | 16.19 | 15.75 | 16.17 | 16.61 | 16.37 |
| daf-21 | 11.88 | 13.46 | 14.18 | 14.07 | 14.54 | 14.81 | 15.13 | 14.96 | 14.93 |
| … | |||||||||
| R11A5.3 | 9.21 | 9.96 | 14.32 | 13.43 | 15.87 | 15.47 | 15.68 | 14.96 | 14.62 |
| dnj-13 | 10.58 | 12.02 | 12.77 | 12.15 | 12.43 | 12.56 | 11.63 | 11.35 | 11.39 |
| unc-23 | 9.66 | 11.09 | 11.99 | 11.42 | 12.19 | 12.32 | 11.48 | 11.70 | 11.51 |
| dnj-12 | 7.43 | 8.60 | 8.44 | 8.49 | 8.83 | 9.04 | 9.01 | 9.36 | 9.31 |
| hsp-1 | 15.13 | 15.71 | 16.56 | 15.87 | 15.70 | 16.03 | 15.60 | 15.30 | 15.16 |
Individual induced genes are shown for 30 min, and 720 min of heat-shock compared to their non-stressed state.
The isolated DBD of HSF-1 shows affinity to the F44E5.4 inducible promoter
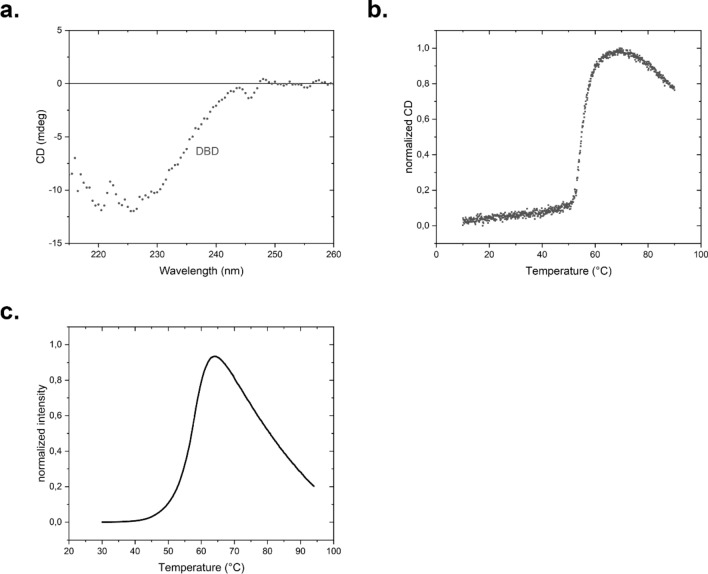
To test, to what extent binding differences correlate with expression differences and structural differences of the HSE we set out to determine in vitro, how the interaction of HSF-1 DBD is at these differently structured HSEs. To this end the isolated DNA binding domain of nematode HSF-1 was purified, containing the DBD and omitting the nematode-specific sequences at the N-terminus and the further regulatory domains at the C-terminus. The structure of the purified DNA-binding domain was investigated by far-UV CD-spectroscopy. The spectra revealed a mostly α-helical structure (Fig. 2a). To confirm the stability of the domain, we performed a thermal transition in the Far-UV CD-range and obtained a temperature midpoint of the unfolding transition of 55 °C (Fig. 2b). We also performed a stability investigation employing the TSA assay, where no obvious differences were observed regarding the melting point (Fig. 2c). Thus, all spectroscopic methods imply that the isolated DNA-binding domain of C. elegans HSF-1 is a stable and structured protein.
Figure 2.
Structure and stability of HSF-1 DBD fragment 82–198. (a) CD-spectroscopy, (b) unfolding, determined by a thermal transition with CD-spectroscopy and (c) unfolding, determined by a thermal shift assay.
dsDNA probes were then generated by us from the heat-shock responsive cluster, in order to gain a better insight into the differential expression form the chaperone-gene derived HSEs. F44E5.4 features a high consensus score pentameric site, both hsp-70 and unc-23 consist of only one trimeric site, while hsp-16.2 has a high consensus score tetrameric site plus an additional trimeric site. Probes of equal length were also made for hsp-1, dnj12 (trimeric HSE-site) and dnj-13 (tetrameric site) representing the non-induced heat-shock related proteins. Since both sequence and position in the promoter region of the following genes are identical the probe for F44E5.4 also represents F44E5.5, while hsp-16.2 represents hsp-16.11, hsp-16.41, hsp-16.48. The sequences of the probes were obtained from the respective promoter regions. Here only HSEs were considered that locate within 1000 bps upstream of the starting point of transcription (Table 6). F44E5.4p contains more HSEs in its sequence than synthesized in this study (comparison of the promoter regions), but here likewise the probes with the highest consensus score were synthesized.
Table 6.
HSE-containing probes designed from the promoter sequences of chaperone genes and used in the binding studies.
| Fwd | Rev | Trimeric consensus scores | Orien-tation | Start position relative to ATG | Promoter location | ORF location, orientation | |||
|---|---|---|---|---|---|---|---|---|---|
| DNJ-12 | aaaagtgtcgagaatgttcacgaaaaatcgttaga | tctaacgatttttcgtgaacattctcgacactttt | 9.66 | Fwd | − 221 | chrI, 14,756,377–14,757,376 | 14,756,377 | 14,757,376 | chrI, 14,756,842–14,758,772 |
| DNJ-13 | agtaaatagaacgctctggaaagttccgcactctt | aagagtgcggaactttccagagcgttctatttact | 13.01, | Fwd, | − 205 | chrII, 11,544,000–11,547,000 | chrII, 11,545,609–11,547,273 | ||
| 9.1 | Rev | + | |||||||
| F44E5.4 (also F44E5.5) | gcagtggaatattccagaagcttctagaagaagtt | aacttcttctagaagcttctggaatattccactgc | 11.59, | Fwd, Rev, Fwd | − 124 | chrII, 11,757,500–11,760,300 | 11,757,016–11,761,764 | ||
| 12.43, | + ,– | ||||||||
| 13.07 | |||||||||
| HSP-1 | tgacgaaattcgaattttctagaatcccgccacgc | gcgtggcgggattctagaaaattcgaatttcgtca | 10.24 | Fwd | − 150 | chrIV, 17,277,000–17,281,000 | chrIV, 17,278,910–17,281,289 | ||
| Hsp-16.2a | gccttacagaatgttctagaaggtcctagatgcat | atgcatctaggaccttctagaacattctgtaaggc | 9.41, | Rev, | − 117 (hsp-16.49 and hsp-16.41) | chrV, 1,804,000–1,806,000 | chrV, 1,804,268–1,804,799 | ||
| 12.81 | Fwd | − 253 (hsp-16.11 and hsp-16.2) | – | ||||||
| Hsp-16.2b | acaagcagctcgaatgttctagaaaaaggtggaaa | tttccacctttttctagaacattcgagctgcttgt | 11.86 | Fwd | − 104 | chrV, 1,804,000–1,806,000 | chrV, 1,804,268–1,804,799 | ||
| – | |||||||||
| Hsp-70 | agtaaattgtagaaggttctagaagatgccagagg | cctctggcatcttctagaaccttctacaatttact | 12.71 | Fwd | − 112 | chrI, 9,319,000–9,323,000 | chrI, 9,320,350–9,322,501 | ||
| – | |||||||||
| Unc-23 | acggagcctcggattcttccagaaaattgagtctc | gagactcaattttctggaagaatccgaggctccgt | 9.48 | Fwd | −573 | chrV, 8,935,000–8,939,000 | chrV, 8,936,195–8,939,472 | ||
| + , + , + | |||||||||
The designed probes originate from the 1000 bp promoter sequence and are positioned as indicated. The strand and anti-sense strand were synthesized and combined to give the promoter sequence able to bind HSEs.
EMSA-assays imply differences between the chaperone-gene derived HSEs
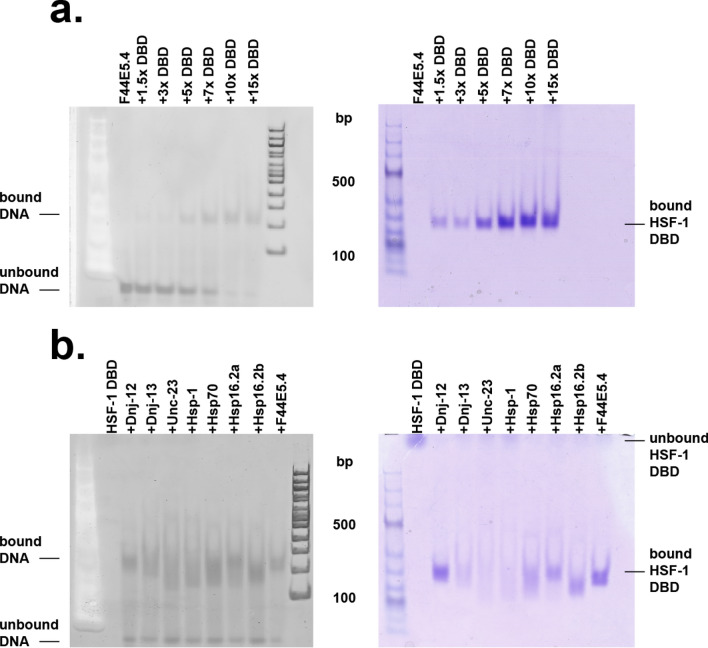
Electrophoretic-mobility shift assays (EMSA) were performed to test the interaction between purified HSF-1 DBD and dsDNAs (Fig. 3a). We set out to perform an initial binding analysis HSF-1 DBD to the promotor of F44E5.4, which also contains the highest amount of HSEs compared to the promoters used in this study. To this end, we titrated the DBD of HSF-1 with concentrations ranging from 0–22.5 µM to 1.5 µM of promoter DNA, which represents a 15-fold molar excess at the highest concentration. Notably a saturated complex of protein and DNA was reached at a concentration of 10 µM HSF1-DBD, at which the complex bands could be observed on the Coomassie stained gel, while at the same time no further reduction in free DNA was visible. Following this initial analysis, we also tested the dsDNA probes of hsp-70, hsp-1, hsp-16.2, unc-23, dnj-12 and dnj-13 under the same conditions. 10 µM HSF-1 DBD was added to each probe to determine the formation of the respective protein-DNA complex (Fig. 3b), which showed depending on the probe used, a highly variable reduction in migration speed. While probes derived from the promoter of dnj-13, unc-23 and hsp-1 hardly showed any interaction with the DBD of HSF-1, F44E5.4, hsp-70, hsp16.2 and dnj-12 derived probes appeared to interact strongly, thereby forming intense bands with HSF-1 DBD, representing the dsDNA-protein complex. These results indicate that the HSF-1 DBD alone can interact with the different promoter-derived HSEs to a different extent.
Figure 3.
EMSA shift of the DNA-HSF-1 complex. The DNAs were stained with the DNA stain (left gel) whereas proteins were stained with Coomassie Blue (right gel). DNA-DBD binding occurs when both stained DNA and stained protein overlap, in comparison to each other. (a) Titration of HSF-1 DBD to the promotor F44E5.4, ranging from a 1,5–15-fold excess of HSF-1 DBD; (b) Comparison of selected DNA promotor sequences, each added to the HSF-1 DBD.
Analytical ultracentrifugation confirms the binding differences at the various HSE-sites
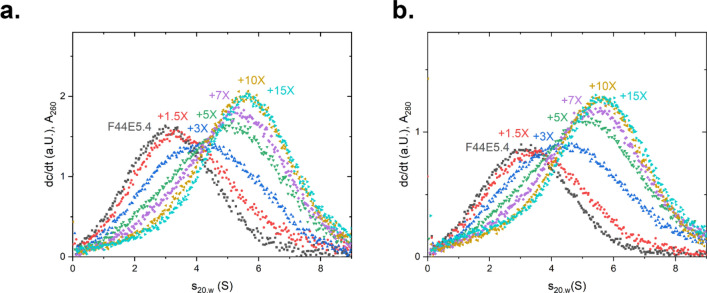
To unravel the interaction patterns, we performed SV-AUC under the condition employed for the gel-based assay. To this end, a titration with the DNA probe representing F44E5.4p was performed. Addition of HSF-1 DBD resulted in an increase in the sedimentation coefficient, indicating the binding of HSF-1 DBD to dsDNA (Fig. 4). In the titration, the progressive binding of HSF-1 DBD molecules increases the s20,w of the main species and indicates further complex formation at higher protein:DNA ratios. The complex with F44E5.4p appears to reach a saturated level when a tenfold excess of HSF-1 DBD is added. At this point, the presence of remaining unbound HSF-1 DBD becomes visible, which is in agreement to the EMSA binding assay.
Figure 4.
Analysis of interaction between promotor F44E5.4 and Hsf-1 DBD via titration in SV-AUC. HSF-1 DBD was titrated to the promoter F44E5.4 in concentrations ranging from a 0–15-fold excess. A shift to the right represents DNA-binding by the HSF-1 DBD. (a) dc/dt plot of the absorbance measured at 260 nm and for (b) at 280 nm at different concentrations of HSF-1 DBD, when added to 1.5 µM of HSE-containing DNA.
Having investigated the promoter region with 5 potential binding sites, we tested, whether the promoter regions with less binding sites, show a similar response. Thus, the same approach was chosen for a DNA with only 3 binding sites derived from the promoter of hsp-70. Here the saturation point of the binding reaction was shifted to lower s20,w values in both wavelength detection modes, suggesting that in this case less HSF-1 DBD molecules bind to the promoter (Fig. 5a). This behavior therefore appears to be a sequence-specific property. Further analog experiments were performed with all the other dsDNA strands and initially the highest s20,w values were noted (Fig. 5b-g).
Figure 5.
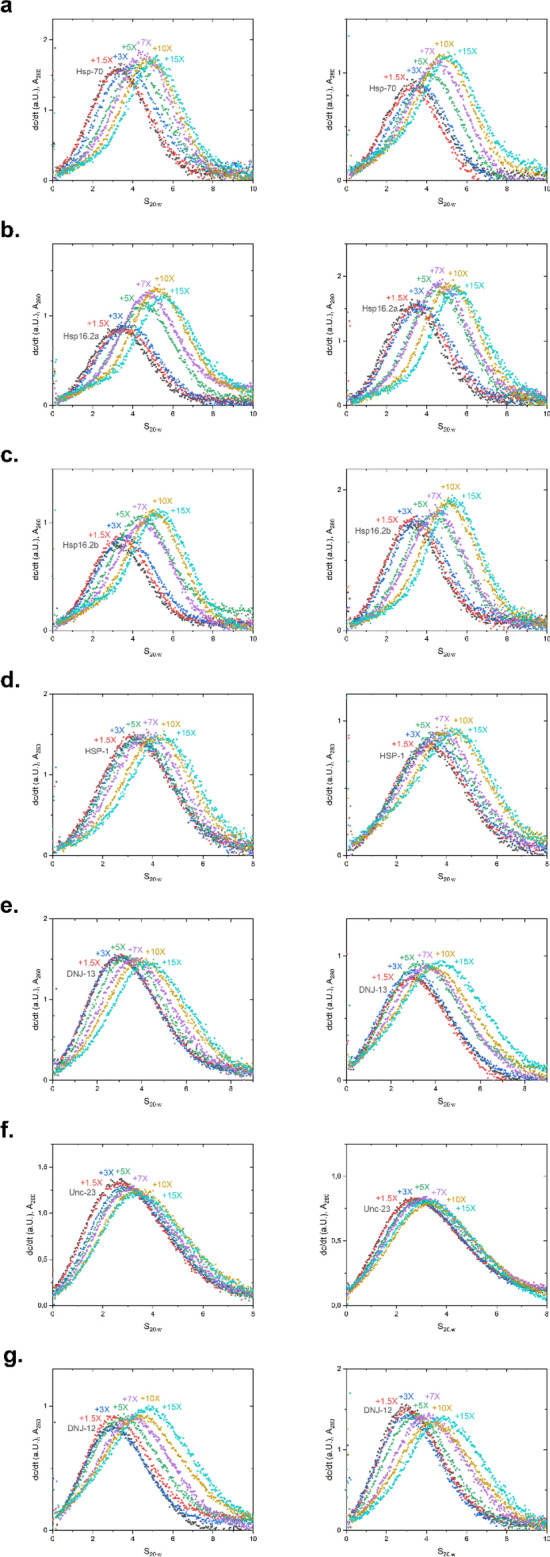
Analysis of interaction between selected promotors and Hsf-1 DBD via titration in SV-AUC. HSF-1 DBD was titrated to each promoter in concentrations ranging from a 0–15-fold excess. A shift to the right represents DNA-binding by the HSF-1 DBD. dc/dt plots of the absorbance measured at 260 nm (left panel) and at 280 nm (right panel). Respective promoters used: (a) HSP-70; (b) HSP16.2a; (c) HSP16.2 b; (d) HSP-1; (e) DNJ-13; (f) UNC-23; (g) DNJ-12.
SV-AUC fitting to defined species reveals potential differences in occupation of complex binding sites
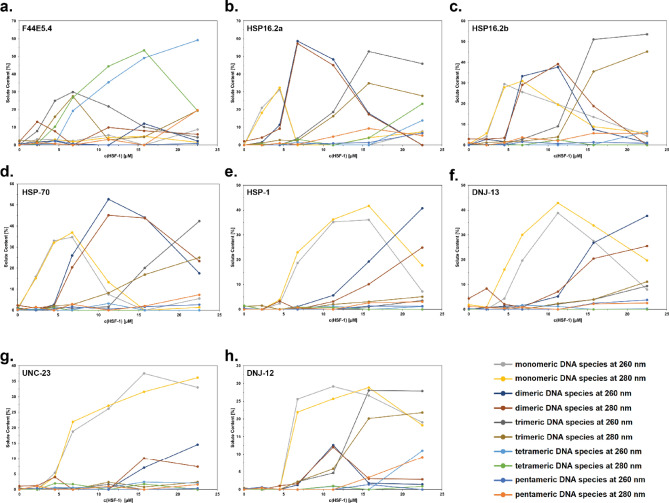
The very weak interaction at several consistent—at least on a monomeric level—HSE sites, questions the independent interaction of monomeric units at these sites. UltraScan III was employed to analyze the data from these experiments and to obtain information on the binding equilibrium in solution. To this end we compared the general ability to fit the data with a very flexible model (2DSA-IT) and with a very constrained model, where a custom grid was designed containing one s20,w value (Table 7) for each species to be considered (2DSA-CG-IT). This method reveals available free protein concentration dependent changes in complex species distributions and offers the opportunity to fit distributions of DNA/protein complex obtained directly from raw data to hypothetical species, thus, to obtain the concentration of each potential complex species and to describe the composition of the complex mixture in each sample. The comparison of RMSD values from the 2DSA-CG-IT fit of each complex species formed with different DNAs is shown in Fig. 6. It is very clear from these data that different assembly mechanisms are happening in different probes and different stoichiometries must be assumed. In the UltraScan III analysis, the higher order complexes are only populated when using larger HSEs and in all cases the buildup of the free HSF-1 DBD can be observed at the higher concentrations employed in each titration. Furthermore, almost no binding was observed for the constructs of hsp-1, dnj-13, and unc-23. (Fig. 6e, f and g).
Table 7.
Parameters used for custom grid fitting approach.
| s20,w (S) | E260 (M−1 cm−1) | E280 (M−1 cm−1) | |
|---|---|---|---|
| HSF-1 DBD | 1.75 | 8,388 | 13,980 |
| dsDNA | 3.15 | 563,198 | 304,431 |
| ssDNA (residual from mixing) | 1.75 | 355,400 | 192,108 |
| dsDNA + 1 DBD | 3.9 | 571,586 | 318,411 |
| dsDNA + 2 DBD | 4.65 | 579,974 | 332,391 |
| dsDNA + 3 DBD | 5.35 | 588,362 | 346,371 |
| dsDNA + 4 DBD | 5.95 | 596,750 | 360,351 |
| dsDNA + 5 DBD | 6.55 | 605,138 | 374,331 |
Generally applicable sedimentation coefficients were obtained for this model system, where HSEs are embedded into probes of identical size. Extinction coefficients are also shown. f/f0 was kept floating during the fit with the custom grid model, while the s20,w was set to allow only the species mentioned in this table.
Figure 6.
Partial concentration of each species in DNA-HSF-1 samples, derived via custom grid fitting. Selected promoter = (a) F44E5.4; (b) HSP16.2a; (c) HSP16.2b; (d) HSP-70; (e) HSP-1; (f) DNJ-13; (g) UNC-23; (h) DNJ-12.
Global fitting of stepwise binding models implies favorable cooperative action at second and third binding steps
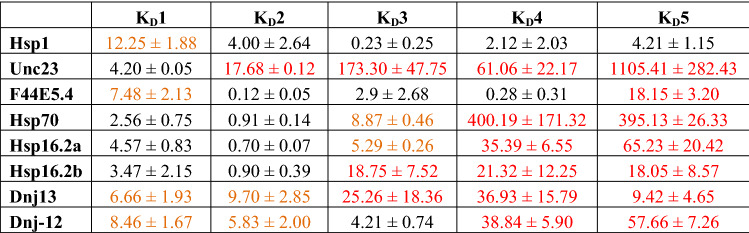
We then set out to globally fit one titration to a predefined set of species, which is kept invariant throughout all the DNA probes analyzed. This is possible, as the dsDNA strands are of equal length and the binding sites are engineered to be in the middle of each dsDNA scaffold. Indeed, for each of the stronger binding species, the second binding step is exposing a lower dissociation constant compared to the first binding steps and similar relationships occur at the later binding steps at probes that harbor more than three binding sites. In fact, the four strongly interacting systems (hsp-70, hsp-16.2a, hsp-16.2b and F44E5.4) show a second binding step with submicromolar affinity, while the first binding step is weaker (Table 8). Thus, it is indeed to be expected that cooperative actions increase the binding affinity and interactions between the occupied binding sites modulate and potentially coordinate the binding of HSF-1 at these HSEs.
Table 8.
Calculated KD values derived from SV-AUC fitting.
No color coding = cooperativity, orange color coding = weak cooperativity, red color coding = no cooperativity.
Discussion
In the nematode genome there are 4120 HSEs, which contain HSF-1 binding consensus regions in the 500 bp upstream of their start codon. It is very surprising that despite these many HSF-1 regulated genes the canonical heat-shock response only represents a clique of 8 genes, 7 of which are regulated by HSF-1 binding promoter regions. Thus, the extent of regulation resulting from HSF-1’s actions is well beyond the induction of stress genes under stress conditions and reaches far into the normal growth cycle of the nematode under non-stressed conditions. The ability to resolve the clique membership based on coexpression analysis shows that also in larger organisms this approach may be successful and able to connect different cliques to different tissues and developmental states.
Binding affinity, cooperativity and stoichiometry on complex promoter sequences
We here tested the binding of the HSF-1 DBD to some of the likely interacting promoter regions. From these studies we can find that the HSF-1 DBD alone can bind the HSE-regions originated from the genome with certain selectivity based on its affinity. Despite this, the affinities correlate to some extent with the calculated consensus score and with the inducibility of the respective gene. It is interesting to note, that despite the proposed trimeric binding mode, tetrameric and pentameric HSEs exist and that binding to those sites is driven by additional cooperativity. Among the probes we investigate in this study, the tetrameric and pentameric sites represent those, which are inducible upon heat-shock.
In general, the developed AUC assay to test the binding of several proteins to one DNA strand is very valuable in quantifying the binding events and may represent an opportunity to study the many interactions occurring on dsDNA with different binding sites for individual transcription factors. While the sedimentation coefficients for the custom grid are an assumption, they provide a rational to obtain stepwise binding information from the SV-AUC titration data. The absolute values of the obtained stepwise dissociation constants are to be used with care, but trends can be derived from these values with good confidence. The ability to resolve different intermediate assembly steps may be further increased by using direct interaction models for the fitting, but the stepwise procedure shown here already represents the chance to quantify these events. Nevertheless, the grouping of the genes into coexpression cliques, the identification of common transcription factors for these cliques and the analysis of binding events to the predicted transcription factor binding sites opens possibilities to gain further insight into the complex relationships leading to the spatio-temporal expression of genes during development and aging of C. elegans, or complex multi-step binding reactions in general.
Correlation between binding and inducibility
Comparing the binding ability of HSF-1 to the promoter regions and the observed response to heat-stress may be far fetching, given that only the DBD of HSF-1 was studied and further regulation will surely come from the other regions of this complex protein. Nevertheless, for the strongest inducible genes, also the highest affinities are observed (hsp-16.2, F44E4.5, hsp-70), which are also in accordance with previous studies15,17.
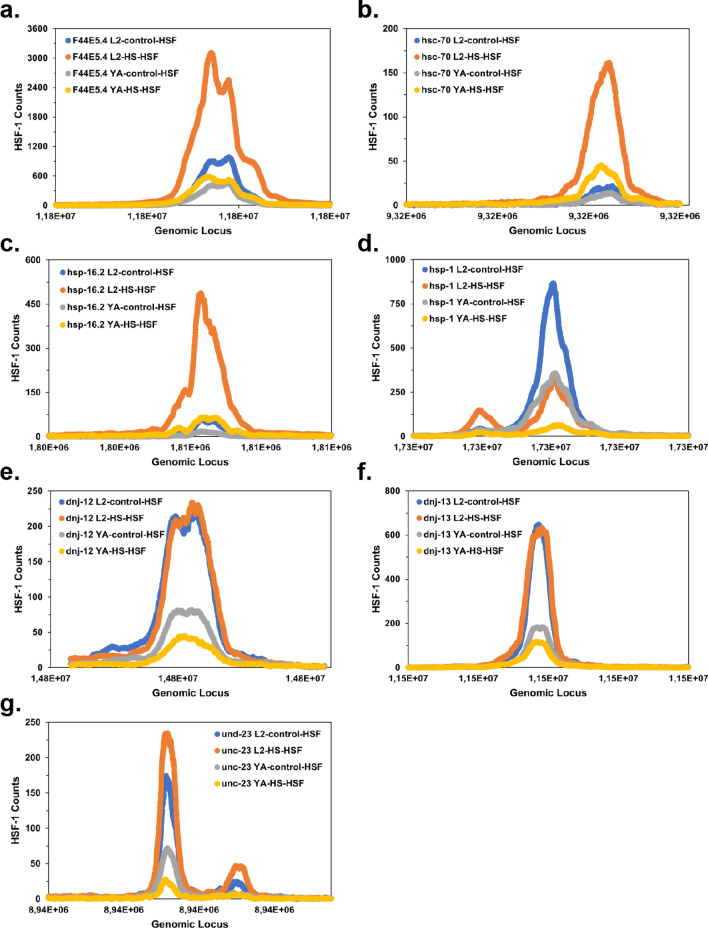
One exception among the probes studied here is dnj-12, which is only weakly inducible but well capable of binding to the HSF-1 DBD. Interestingly dnj-12 is already at non-stressed conditions highly expressed, similarly to hsp-1. This can be derived from the relatively high number of RNAseq reads originating from these ORFs. Given the ubiquitous expression of this protein it might be envisioned that its binding to HSF-1 is constitutive, and the induced expression therefore is not increased upon heat-shock. Looking into publicly available ChIPseq data17 for the locations described here, some of these speculations can be tested. Indeed, for the genes coexpressed upon heat-shock, hsp-16, F44E5.4 and hsp-70 this can be confirmed (Fig. 7) and the inducibility from the promoters F44E5.4/5, hsp-16.2 and hsp-70 correlates well with increased occupancy of HSF-1 on the HSE-sites. Even for unc-23 a slight increase in occupancy can be observed. This change at the promoter regions cannot be observed for the non-inducible probes. Here (dnj-12, dnj-13 and hsp-1), HSF-1 sites are occupied in a similar or even reduced manner with and without heat-shock implying a constitutive expression and possible constitutive function of HSF-1 responsible for the high expression levels observed for these genes under stressed and non-stressed conditions. This logic may be relevant for several of the 4120 HSE-binding sites found in promoter regions. Despite the correlations observed, it is important to note, that the approach employed in this study solely considers the DBD of HSF-1 and that HSF-1 HSE binding in the cell is further regulated by other regulatory domains, oligomerization domains and posttranslational modifications, like phosphorylation39,40 and deacetylation41. Due to these limitations further studies with longer fragments or full-length protein will need to be performed to unravel the full relationship between promoter sequences and HSF-1 binding.
Figure 7.
Promoter regions of C. elegans were investigated to identify the occupancy as determined by HSF-1 ChIPseq data of Li et al.17. Four experiments were compared based on the available data: Young adult with and without heat-shock and L2 larvae with and without heat-shock for all the promoter regions investigated here.
Therefore, the here applied approach shows the direct affinity of the unmodified DBD to the DNA, but will require adaptations, when used for the dsDNA binding analysis of full-length HSF-1 in the future.
Supplementary Information
Acknowledgements
K.R., L.S. and S.S. thank the Deutsche Forschungsgemeinschaft for funding the research grant RI1873/1-4 and the Heisenberg position RI1873/5-1. We also thank the distributors of free and open-source software that we could use during this study. This includes UltraScan III Version 4.0 (https://ultrascan3.aucsolutions.com/), RMAexpress 1.0.5 by Ben Bolstad (https://rmaexpress.bmbolstad.com/), Cytoscape 3.8.2 (Cytoscape Consortium, https://cytoscape.org/) and Subio64 software package 1.24.5853 (Subio Inc. Kagoshima, Japan).
Author contributions
S.S. and K.R. designed the experiments, S.S., L.S., A.B.H., R.C., performed the experiments, S.S., L.S. and K.R. analyzed the data and wrote the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All data are fully available without restriction.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-12736-x.
References
- 1.Vihervaara A, Sistonen L. HSF1 at a glance. J. Cell Sci. 2014;127:261–266. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 2.Skaggs HS, et al. HSF1-TPR interaction facilitates export of stress-induced HSP70 mRNA. J. Biol. Chem. 2007;282:33902–33907. doi: 10.1074/jbc.M704054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams RS, Benjamin IJ. Protective responses in the ischemic myocardium. J. Clin. Invest. 2000;106:813–818. doi: 10.1172/JCI11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao X, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kallio M, et al. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21:2591–2601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaitsuka T, Tomizawa K, Matsushita M. Transformation of eEF1Bdelta into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011;12:673–681. doi: 10.1038/embor.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubota H, Matsumoto S, Yokota S, Yanagi H, Yura T. Transcriptional activation of mouse cytosolic chaperonin CCT subunit genes by heat shock factors HSF1 and HSF2. FEBS Lett. 1999;461:125–129. doi: 10.1016/s0014-5793(99)01437-4. [DOI] [PubMed] [Google Scholar]
- 8.Cahill CM, Waterman WR, Xie Y, Auron PE, Calderwood SK. Transcriptional repression of the prointerleukin 1beta gene by heat shock factor 1. J. Biol. Chem. 1996;271:24874–24879. doi: 10.1016/S0021-9258(18)40087-7. [DOI] [PubMed] [Google Scholar]
- 9.Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 10.Gidalevitz, T., Prahlad, V., & Morimoto, R. I. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb Perspect Biol10.1101/cshperspect.a009704 (2011). [DOI] [PMC free article] [PubMed]
- 11.Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010;6:e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendillo ML, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol. 2017;27:895–905. doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joutsen, J. & Sistonen, L. Tailoring of proteostasis networks with heat shock factors. Cold Spring Harb Perspect Biol11, doi:10.1101/cshperspect.a034066 (2019). [DOI] [PMC free article] [PubMed]
- 15.Brunquell J, Morris S, Lu Y, Cheng F, Westerheide SD. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics. 2016;17:559. doi: 10.1186/s12864-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 2013;9:e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Chauve L, Phelps G, Brielmann RM, Morimoto RI. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 2016;30:2062–2075. doi: 10.1101/gad.283317.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbadia J, Morimoto RI. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 20.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.e03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunquell J, Snyder A, Cheng F, Westerheide SD. HSF-1 is a regulator of miRNA expression in Caenorhabditis elegans. PLoS ONE. 2017;12:e0183445. doi: 10.1371/journal.pone.0183445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp. Quant. Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 24.GuhaThakurta D, et al. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 2002;12:701–712. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaiser AM, Kaiser CJ, Haslbeck V, Richter K. Downregulation of the Hsp90 system causes defects in muscle cells of Caenorhabditis elegans. PLoS ONE. 2011;6:e25485. doi: 10.1371/journal.pone.0025485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, et al. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. J. Biol. Chem. 2006;281:782–791. doi: 10.1074/jbc.M505822200. [DOI] [PubMed] [Google Scholar]
- 27.Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J. Biol. Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
- 28.Guo Y, et al. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J. Biol. Chem. 2001;276:45791–45799. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- 29.Sima S, Schmauder L, Richter K. Genome-wide analysis of yeast expression data based on a priori generated co-regulation cliques. Microb. Cell. 2019;6:160–176. doi: 10.15698/mic2019.03.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmauder, L., Richter, K. hsp-90 and unc-45 depletion induce characteristic transcriptional signatures in coexpression cliques of C elegans. Sci Rep11, 12852, 10.1038/s41598-021-91690-6 (2021). [DOI] [PMC free article] [PubMed]
- 31.Wang P, Zhao J, Corsi AK. Identification of novel target genes of CeTwist and CeE/DA. Dev. Biol. 2006;293:486–498. doi: 10.1016/j.ydbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Jovic K, et al. Temporal dynamics of gene expression in heat-stressed Caenorhabditis elegans. PLoS ONE. 2017;12:e0189445. doi: 10.1371/journal.pone.0189445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Cheng X, Li Y, Wu M, Zhao Y. Image-based promoter prediction: a promoter prediction method based on evolutionarily generated patterns. Sci. Rep. 2018;8:17695. doi: 10.1038/s41598-018-36308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demeler B, Brookes E, Nagel-Steger L. Analysis of heterogeneity in molecular weight and shape by analytical ultracentrifugation using parallel distributed computing. Methods Enzymol. 2009;454:87–113. doi: 10.1016/S0076-6879(08)03804-4. [DOI] [PubMed] [Google Scholar]
- 37.Haslbeck V, et al. The activity of protein phosphatase 5 towards native clients is modulated by the middle- and C-terminal domains of Hsp90. Sci. Rep. 2015;5:17058. doi: 10.1038/srep17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrigues, J. M., Tsu, B. V., Daugherty, M. D. & Pasquinelli, A. E. Diversification of the Caenorhabditis heat shock response by Helitron transposable elements. eLife8, e51139, doi:10.7554/eLife.51139 (2019). [DOI] [PMC free article] [PubMed]
- 39.Hietakangas V, et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmberg CI, Tran SE, Eriksson JE, Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 41.Zelin E, Freeman BC. Lysine deacetylases regulate the heat shock response including the age-associated impairment of HSF1. J. Mol. Biol. 2015;427:1644–1654. doi: 10.1016/j.jmb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are fully available without restriction.