Abstract
Microbial natural products have provided an important source of therapeutic leads and motivated research and innovation in diverse scientific disciplines. In recent years, it has become evident that bacteria harbor a large, hidden reservoir of potential natural products in the form of silent or cryptic biosynthetic gene clusters (BGCs). These can be readily identified in microbial genome sequences but do not give rise to detectable levels of a natural product. Herein, we provide a useful organizational framework for the various methods that have been implemented for interrogating silent BGCs. We divide all available approaches into four categories. The first three are endogenous strategies that utilize the native host in conjunction with classical genetics, chemical genetics, or different culture modalities. The last category comprises expression of the entire BGC in a heterologous host. For each category, we describe the rationale, recent applications, and associated advantages and limitations.
Keywords: natural product, bacteria, biosynthesis, silent gene cluster, cryptic metabolite
INTRODUCTION
Virtually all microbes use small molecules for nutrient acquisition, communication, and perhaps most importantly, chemical warfare against competing neighbors. The discovery of and further investigation into these primarily extracellular molecules, termed natural products (NPs) or secondary metabolites (SMs), provided the foundations for modern medicine (1). The clinical deployment of penicillin, a fungal NP, revolutionized medicine during a period when the top three leading causes of death were infectious disease. Since then, many US Food and Drug Administration (FDA)-approved drugs have been derived from NPs, and, despite decades of mining, NPs remain an important source for drug discovery (2).
NPs are typically biosynthesized by multiple enzymes, and the genes encoding them are often clustered in microbial genomes. Referred to as biosynthetic gene clusters (BGCs), the sequence signatures of well-known enzymes can now be readily identified using a variety of publicly available programs such as antiSMASH (3) or PRISM (4). This type of analysis with available microbial genomes shows that the products of most identifiable BGCs remain to be determined, even in familiar strains (5). Thus, our arsenal of natural therapeutic agents is based on a small fraction of microbial biosynthetic capacity, and methods that unlock the remaining majority could have a significant impact on NP research and, thereby, on drug discovery.
Several criteria must be met for the product of a given BGC to be detected. The BGC of interest must be sufficiently transcribed and translated. The active biosynthetic enzymes must then have at their disposal all necessary cofactors and substrates needed to synthesize the target NP. As there is no universal method capable of extracting all metabolites, the appropriate extraction method must be used. In addition, the desired NP must also be synthesized above the biological or instrumental limits of detection for isolation and characterization. Based on genomic evidence, one or more of these conditions is unsatisfied for the majority of BGCs, leading to a discrepancy between biosynthetic potential and measurable NP output when bacteria are interrogated in the laboratory. These BGCs are referred to as orphan, cryptic, or silent (see the sidebar titled Biosynthetic Gene Clusters: Constitutive, Silent, Cryptic, or Orphan?).
While the prospect of a so-far hidden source of NPs is exciting, accessing these cryptic NPs is challenging, so much so that many investigators have instead shifted their attention to new NP sources, such as the human microbiome (6, 7), marine (8, 9) and hypogean (10, 11) environments, uncultured strains (12), and other underexplored microbial phyla. Mining new and understudied microbial lineages has proven successful in identifying novel NPs. However, cryptic BGCs are just as common in these bacteria, and overcoming the multifaceted challenge of identifying their products is therefore likely to remain a focal point for NP research for years to come. Herein, we describe and evaluate the advantages and limitations of current approaches for accessing the products of silent BGCs (see the sidebar titled How Much Expression Is Enough?). We divide the available techniques into four categories: genetics-reliant methods, genetics-independent (or chemical-genetic) methods, culture modalities, and heterologous expression (Figure 1). The first three are endogenous strategies using the native producer, while the latter occurs exogenously. We hope this review provides a useful organizing principle for extant methodologies and serves as a guide to investigators interested in activating silent BGCs in diverse bacteria.
Figure 1.
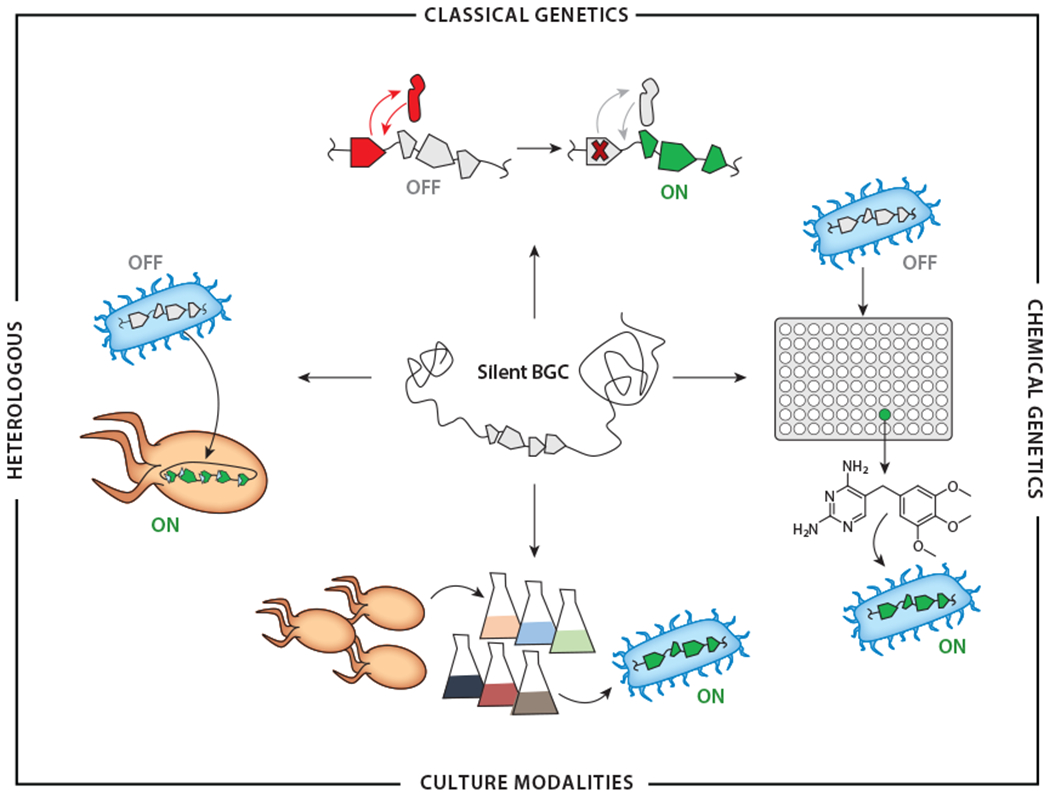
Categories of approaches to activate silent BGCs. In classical genetics (top), forward or reverse genetic manipulations are used to enhance expression of silent BGCs. These can include deletion of transcriptional repressors, as depicted. In chemical genetics (right), libraries of small molecules provide candidate elicitors of silent BGCs, as shown with the example of trimethoprim. Alternatively, the activity of relevant pathways or proteins can be modulated with specific small molecules in reverse chemical-genetic applications. Different culture modalities (bottom), in either mono- or mixed-culture fermentation, can also lead to induction of sparingly expressed BGCs. In heterologous expression (left), the entire BGC is transplanted into a desired host with regulatory elements designed to gain access to the products of a silent BGC. Abbreviation: BGC, biosynthetic gene cluster.
ENDOGENOUS VERSUS EXOGENOUS ACTIVATION OF SILENT BIOSYNTHETIC GENE CLUSTERS
The first important distinction between the various methodologies is using the native host versus a heterologous one for activation of a silent BGC, an endogenous or exogenous approach, respectively (Figure 1). The main advantage of the endogenous approach is physiological relevance. Molecules discovered from the native producer are the intended metabolites, and they quickly lead to broader questions about the molecules’ biosynthesis, biological activity, molecular target, and chemical ecology. A minor drawback of the approach is practicality: Not all native producers can be easily cultured in the laboratory. This is where heterologous expression provides an advantage, as products of difficult-to-grow bacteria, uncultured microbes, and even fragments of DNA (e.g., from environmental samples) can be investigated. Heterologous expression, therefore, opens the door to the products of BGCs that can be spotted in metagenomic DNA sequences. In addition, successful heterologous production of a metabolite simplifies and facilitates efforts to engineer variants, as the mobilized biosynthetic genes can be manipulated. The main disadvantage of this approach is that the relevance of the molecules identified needs to be demonstrated where possible, for example, by detection of the same metabolite in the native producer. An appropriate expression host is critical, and the method is not suitable for metabolites that arise from cross talk between different BGCs or genes encoded in disparate genomic loci; it is also laborious with BGCs composed of multiple operons. Moreover, despite recent successes, there is generally a size limit to heterologous expression, as it remains challenging to reliably package and move large pieces of DNA. Nonetheless, heterologous and endogenous means of production are largely complementary, and the suitability of each for a given BGC needs to be evaluated on a case-by-case basis with the above caveats in mind.
ENDOGENOUS ACTIVATION: CLASSICAL GENETICS
Forward Genetics
In the endogenous category, classical genetics is a commonly used strategy for activating silent BGCs. It can be conveniently subdivided into forward and reverse genetic methods (Figure 2). Reporter-guided mutant selection (RGMS) is the main technique in the forward direction. It consists of two steps: (a) generation of a random mutant library and (b) selection of the mutant that expresses the silent BGC (Figure 2a). Thus far, UV-induced mutagenesis and transposon (Tn) mutagenesis have been used to generate mutant libraries, which have been probed with phenotypic analyses, reporter genes, and more recently, advanced metabolomics (Figure 3).
Figure 2.
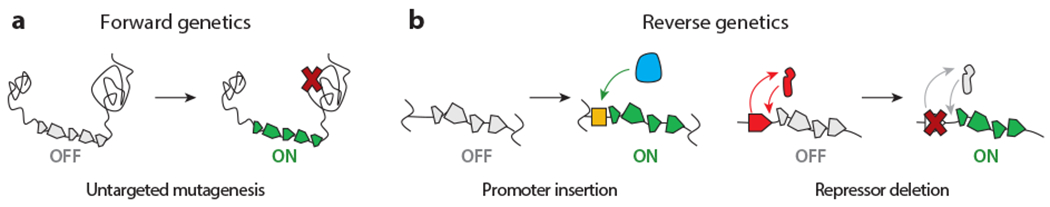
Overview of classical genetic activation approaches. (a) Forward genetics involves untargeted gene mutagenesis, which either directly or indirectly affects the expression of a BGC of interest. (b) Reverse genetic strategies activate cryptic BGCs through targeted gene manipulation, such as promoter insertions (left) or repressor deletions (right). The promoter, RNA polymerase, and repressor are indicated in yellow, blue, and red, respectively. Abbreviation: BGC, biosynthetic gene cluster.
Figure 3.
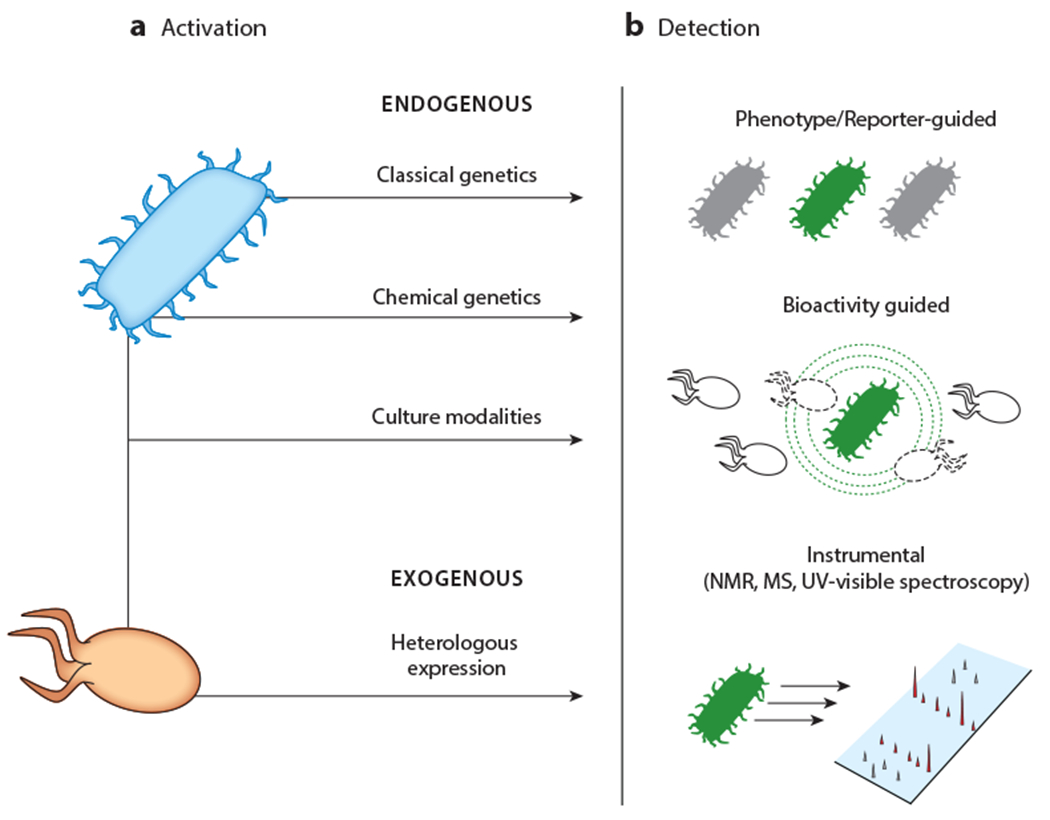
Phases of activation of silent BGCs. Regardless of the method used, the process consists of two steps. (a) A variety of methods can be used for expression of silent BGCs in an endogenous or exogenous host. (b) Upon induction, production of cryptic metabolites can be monitored with several approaches, including visual inspection; genetic reporters; various bioactivity assays including antimicrobial activity; and diverse instrumental methods, such as HPLC-MS, IMS, and UV-visible or NMR spectroscopy. Abbreviations: BGC, biosynthetic gene cluster; HPLC, high-performance liquid chromatography; IMS, imaging mass spectrometry; MS, mass spectrometry; NMR, nuclear magnetic resonance.
The first reports of RGMS focused on increasing the production titers of known metabolites. In the fungus Aspergillus terreus, the method was used to increase yields of the cholesterol-lowering drug lovastatin (14). The promoter of the biosynthetic gene lovF was fused to a phleomycin resistance marker, and a plasmid carrying the construct was transformed into A. terreus. Mutants were generated with UV mutagenesis, and those with greater resistance to phleomycin, likely due to upregulation of the lov BGC, were then examined for lovastatin production. Approximately half the mutants selected exhibited enhanced production. A similar approach was used in Streptomyces clavuligerus to increase yields of clavulanic acid, a clinically used β-lactamase inhibitor. In this case, a double-reporter system was used to lower the rates of false positive hits; 90% of the mutants displayed augmented clavulanic acid titers (15).
With the method established, RGMS was next applied to a silent BGC with an unknown product in Streptomyces sp. PGA64 by Guo et al. (16). A double-reporter construct was employed in which a xylE–neo cassette was fused to the promoter of the silent pga gene cluster. Neo and xylE code for kanamycin acetyl transferase, which confers kanamycin resistance, and a catecholase that stains colonies brown upon catechol treatment, respectively. Upon generation of a mutant library with UV-induced mutagenesis, cells were selected that were catecholase-positive and displayed lowered susceptibility to kanamycin. One of the resulting mutants enabled the identification of the product of the pga locus, leading to the discovery of two novel glycosylated gaudimycin analogs (Figure 4). Additional applications of RGMS in actinomycetes have followed, notably with the inclusion of Tn mutagenesis coupled with either a phenotypic assay in Streptomyces coelicolor (17) or a reporter-gene readout in Streptomyces albus (18). The advantage of Tn mutagenesis is that the gene, the disruption of which (via Tn insertion) leads to activation of the selected BGC, can be easily identified, thus facilitating investigations into the mechanisms underlying induction.
Figure 4.
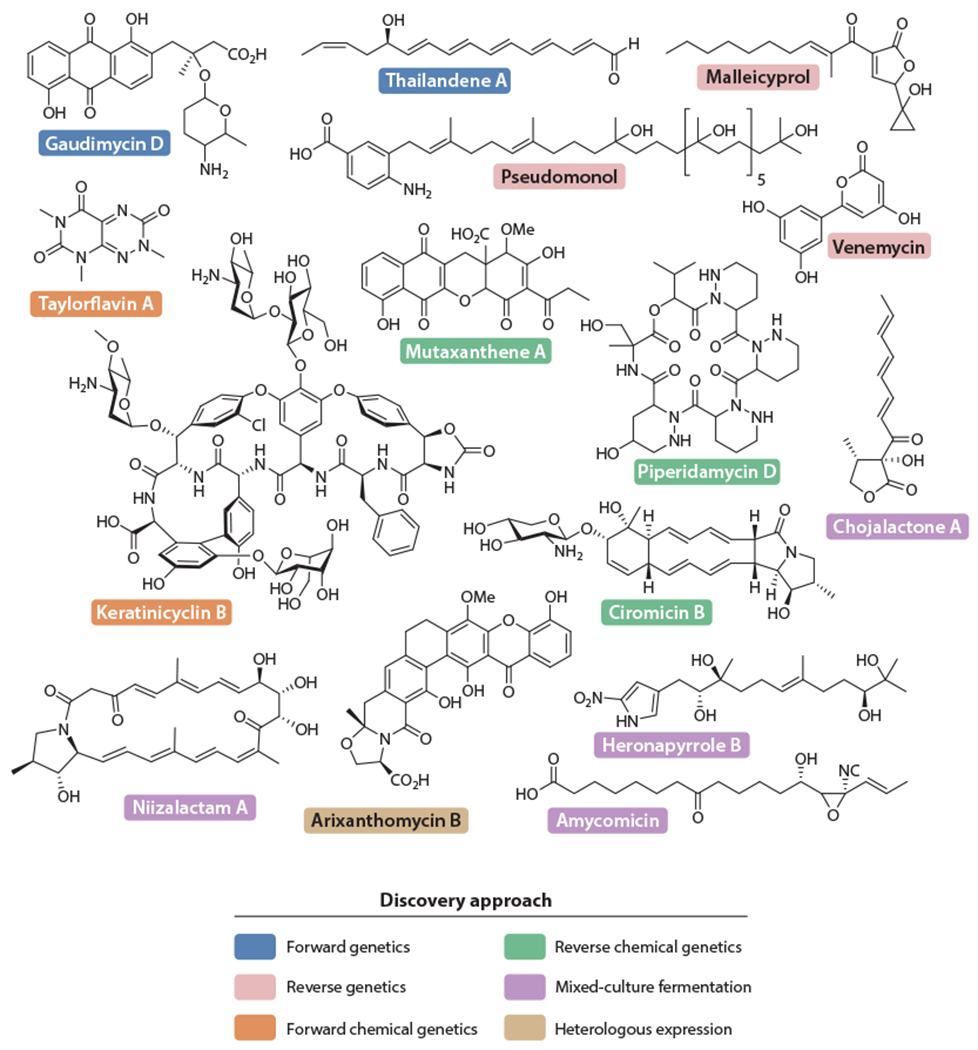
Selected cryptic metabolites discussed in this review. Gaudimycin D and thailandene A (blue) were discovered via RGMS. Pseudomonol, venemycin, and malleicyprol (pink) were identified via applications of reverse genetics. Forward chemical genetic strategies gave rise to keratinicyclin B and taylorflavin A (orange). Reverse chemical genetics led to the discovery of piperidamycin D, mutaxanthene A, and the ciromicins (green). Chojalactone A, niizalactam A, amycomicin, and heronapyrroles (purple) emerged from mixed culture fermentation. Arixanthomycin B (light brown) was found via heterologous expression. Note that ciromicin B was also detected with coculture methods. Abbreviations: BGC, biosynthetic gene cluster; RGMS, reporter-guided mutant selection.
Recently, RGMS has been extended to the Gram-negative Burkholderia genus. In the first rendition, Burkholderia thailandensis was mutagenized with a Tn carrying a gentamicin marker, and the resulting mutants were inspected for phenotypic changes (19). Several mutants exhibited a deep-yellow pigmented phenotype. Transcriptomic and proteomic analyses showed that the expression of a silent iterative type I polyketide synthase had been enhanced. Assessment of these mutants led to the discovery of thailandenes (Figure 4), three novel polyene NPs with antimicrobial activity. More recently, Tn mutagenesis has been combined with genetic reporters or with mass spectrometry (MS)-based detection. In the former case, transcriptional or translational fluorescent protein reporters were generated for three different silent BGCs in B. thailandensis. Subsequent Tn mutagenesis resulted in induction of all three clusters, and follow-up studies showed that interference with pyrimidine nucleotide biosynthesis, via Tn insertion into pyrF, was responsible for induction of the malleicyprol BGC (20). The synthesis of this cytotoxin, therefore, can be triggered by limited nucleotide pools. In the latter case, Tn mutant libraries of Burkholderia plantarii and Burkholderia gladioli were subjected to high-performance liquid chromatography (HPLC)-MS and imaging MS (IMS) analysis, respectively, thereby eliminating genetic manipulations and simultaneously providing a global view of secondary metabolism in response to hundreds of Tn insertions (21). The HPLC-MS data with B. plantarii were assessed using self-organizing map analytics, leading to the discovery of six cryptic metabolites: haereoplantins A–E and burrioplantin (21). IMS of ~1,000 Tn mutants of B. gladioli identified gladiobactin, a cryptic siderophore with a pair of unusual diazonium diolate moieties, which were first described by Hermenau et al. (22).
With several approaches available for generating mutant libraries and detecting SM production, the use of RGMS, or variations thereof, is likely to expand in the future. Importantly, the studies performed so far indicate that even subsaturating libraries are sufficient for significant activation of a silent BGC (17–21). Because RGMS is an endogenous approach that utilizes native regulatory circuits, it has proven useful for investigating both the products and mechanisms underlying the activation of silent BGCs. A drawback is that the microorganism in question needs to be genetically tractable, and this limits the reach of RGMS. But, with the use of UV-induced mutagenesis coupled with genetics-independent readouts, it is conceivable that this technique, or variations thereof, can be applied to genetically intractable strains in the future.
Reverse Genetics
Broadly speaking, two reverse genetic methods have been implemented for activating silent BGCs: (a) modification, overexpression, or deletion of regulatory genes and (b) insertion of constitutively active or inducible promoters to directly drive expression of a BGC (Figure 2b). Other biosynthetic elements, e.g., phosphopantetheinyl transferases, have been exploited as well (23) but so far to a much lesser degree.
Regulatory genes can often be identified within or proximal to a given BGC. Some well-known examples include the γ-butyrolactone receptors (24), which are prevalent in Gram-positive bacteria; LuxR-type regulators (25), which are prevalent in Gram-negative bacteria; and LysR-type transcriptional regulators, which occur in all bacterial phyla (26). Inducing silent BGC regulators, by either introducing constitutive promoters upstream of the regulator or inserting multiple copies of the regulatory gene, has been used to increase production of known metabolites (27–31) and to identify new ones (32–35). For example, a new biaryl polyketide venemycin from Streptomyces venezuelae and the cytotoxic 51-membered glycosylated macrolides stambomycins, encoded by a 150-kb gene cluster in Streptomyces ambofaciens, were discovered when nearby large ATP-binding regulators of the LuxR family (LAL) proteins were constitutively expressed (Figure 4) (32, 33). In these cases, the LAL proteins positively regulate expression by binding to the promoter region(s) upstream and/or within the BGC. There are also many examples of repressors, where inactivation via gene deletion derepresses expression of a BGC and boosts biosynthesis of the cognate NP. The TetR family of transcriptional regulators can be found in some NP BGCs and often maintain such clusters in a silent state under standard growth conditions (36, 37). Inactivation of TetR-type repressors has been associated with the overproduction or induction of several metabolites from otherwise-silent BGCs (38–40). Alternatively, the repressive transcription factors can be sequestered by a plasmid containing multiple copies of the target DNA sequence transformed into the producing bacteria. This transcription factor decoy strategy was recently used to locate a new oxazolomycin analog from Streptomyces sp. NRRL F-4335 (41).
Transcription of bacterial BGCs may be controlled in a pathway-specific manner, as described in the preceding paragraph, and/or by global regulators such as DasR (42), AdpA (43), AbsA2 (44), Lsr2 (45), ScmR (46), or MftR (47). Altering the expression of these pleiotropic regulators may have broader and less predictable effects on BGC activation. For instance, inactivation of AdpA in Streptomyces ansochromogenes abolished production of nikkomycin but activated the production of oviedomycin (48). Similarly, deletion of dasR upregulates many BGCs in S. coelicolor (49) but results in downregulation of secondary metabolism in Saccharopolyspora erythraea (50). Lsr2 was shown to be a global repressor of secondary metabolism, especially for volatile metabolites in S. venezuelae (45), and deletion of scmR and a Campbell insertion into mftR led to broad induction of secondary metabolism in B. thailandensis (46, 47). Modulating the expression of pathway-specific or suspected global regulators is a productive method for altering secondary metabolism and identifying new NPs in genetically tractable hosts. Coupled with MS-based metabolomics, this strategy can provide insights into differential regulation of the various BGCs within a bacterium.
Very recently, a clever strategy to modulate expression of BGCs has been reported in which a hybrid regulatory protein was used (51). LuxR- and LysR-type regulators usually contain a DNA-binding domain with a helix-turn-helix motif and a ligand-binding domain, also referred to as a coinducer-binding domain. A bacterium may not be able to produce the required ligand under standard growth conditions or may rely on other strains for the signal to activate silent BGC(s). Some strains, for example, encode orphan LuxR proteins, a LuxR regulator without the cognate LuxI synthase, which generates the acyl homoserine lactone (AHL) signal that triggers LuxR activity (25). Mukherji et al. (51) replaced an orphan LuxR receptor, which regulates a cryptic BGC in Pseudomonas sp. SZ57, with a chimeric receptor containing the native DNA-binding domain and the ligand-binding domain from LasR, a LuxR protein that binds 3-oxo-decanoyl-homoserine lactone (HSL). When the growth medium was supplemented with this HSL ligand, the activated chimeric LuxR upregulated the target BGC leading to the discovery of a novel, cryptic NP, pseudomonol (Figure 4).
The second broadly used reverse-genetic strategy, which involves replacing the native regulatory elements altogether with an inducible or constitutively active promoter, is the most direct approach for controlling silent BGC expression. Recently, it was applied to S. albus to boost production of 6-epi-alteramides (52). Both replacement of the native promoter and expression of the pathway-specific LuxR family regulator were carried out. Interestingly, enhancing the expression of LuxR resulted in significantly more alteramide production in this case. One advantage of inserting promoters is the added ability to control production of NP variants. For instance, positioning the new promoter to exclude an auxiliary hydroxylase in the alteramide BGC led to high levels of nonhydroxylated 6-epi-alteramide B. The promoter replacement approach has likewise been exploited in genetically tractable proteobacteria, with the discoveries of the cytotoxin malleicyprol from B. thailandensis (53) (Figure 4) and virulence factors from Burkholderia pseudomallei (54) serving as representative examples.
Because genetic manipulation of prolific genera, such as Streptomyces spp. or Amycolatopsis spp., can be laborious and time-consuming, emerging technologies have been adapted to facilitate this process. Notable here is the application of CRISPR–Cas9 methods, a versatile genome-editing technology, which was tailored for streptomycetes by several groups and subsequently used to insert active promoters and thereby drive production of several cryptic metabolites, particularly a novel pigment in Streptomyces viridochromogenes (55).
Reverse-genetic methods are among the most frequently used approaches for inducing silent BGCs. The advantage here is that knowledge regarding the BGC and its regulation can be used to modify the level of expression of the BGC. It also carries the benefits of endogenous approaches mentioned in the section titled Endogenous Versus Exogenous Activation of Silent Biosynthetic Gene Clusters. The main disadvantage is the requirement for genetic tractability. As most microorganisms cannot be genetically manipulated—many cannot even be cultured—the scope of this approach is limited. If a BGC comprises multiple operons, each needs to be induced with a separate promoter. The method is therefore best suited for monooperonic BGCs. In addition, insertion of artificial regulatory elements, such as constitutive promoters, precludes investigations into the native regulatory circuits that turn on silent BGCs, which remains an underexplored area.
ENDOGENOUS ACTIVATION: CHEMICAL GENETICS
Forward Chemical Genetics
Given that genetic manipulations are difficult or impossible in many bacteria, chemical-genetic approaches provide an appealing alternative (Figure 5). Rather than generate Tn mutant libraries, high-throughput small molecule elicitation may be carried out. Rather than delete a given gene, small-molecule inhibitors may be employed. In this section, we cover forward and reverse chemical-genetic methods that have been developed in order to gain insights into cryptic metabolism.
Figure 5.
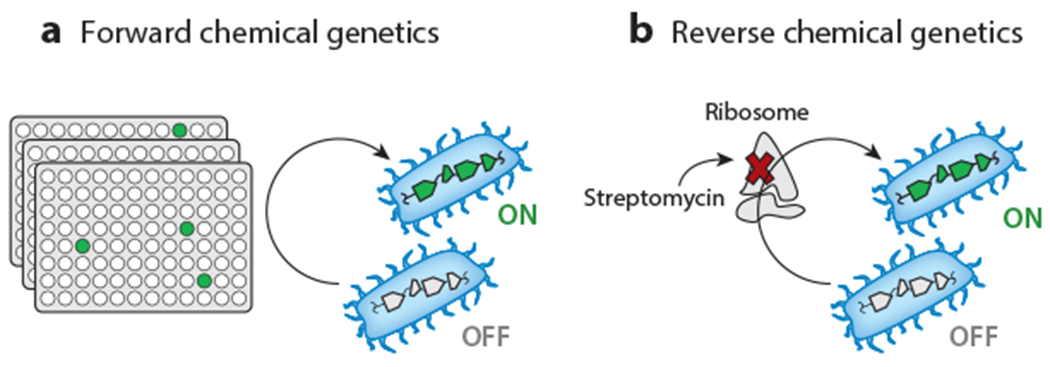
Overview of induction of silent BGCs by chemical genetics. (a) Forward chemical-genetic approaches screen libraries of small molecules to identify elicitors of secondary metabolism, which can be monitored by a variety of methods. (b) Reverse chemical-genetic strategies activate cryptic BGCs by using small molecules to selectively inhibit or alter a chosen target, such as the ribosome by the antibiotic streptomycin. Abbreviation: BGC, biosynthetic gene cluster.
Much like forward genetic approaches to secondary metabolism, forward chemical-genetic approaches also consist of two steps: addition of a library of small molecules to perturb secondary metabolism and detection of that perturbation via a desired readout (Figure 3). The forward chemical-genetic approach has been termed HiTES (high-throughput elicitor screening), as elicitors, i.e., molecules that induce expression of a BGC, are selected from a library of small molecules (Figure 5a). Applications of this method, or variations thereof, were first reported in S. coelicolor and B. thailandensis.
Craney et al. (56) used a large library of synthetic small molecules (>30,000) as candidate elicitors and altered pigmentation as a colorimetric readout to apply the approach to S. coelicolor. The study identified a series of substituted bisaryl ether metabolites, the ARC2 compounds, which induced actinorhodin biosynthesis 2–5-fold depending on the exact elicitor. ARC2 affected secondary metabolism beyond actinorhodin and also altered yields of NPs in other actinomycetes, indicating that its target and mechanism of action were conserved among the strains tested (56).
HiTES in B. thailandensis employed a different source of elicitors and a more general detection method: A library of NPs or clinical agents was used as candidate elicitors, and expression of the silent malleicyprol (mal) BGC, a cytotoxin required for Caenorhabditis elegans infection, was monitored via a mal–lacZ reporter (57). Surprisingly, low-dose antibiotics, including trimethoprim and piperacillin, were identified as the best elicitors of the mal BGC. These antibiotics kill B. thailandensis at high concentrations; at subinhibitory concentrations, however, they induced secondary metabolism (57, 58). Trimethoprim was subsequently shown to serve as a global regulator of secondary metabolism in B. thailandensis, as it induced 21 of 22 BGCs (59, 60). It similarly induced the mal BGC in the related pathogen B. pseudomallei (61). In another HiTES application, the silent sur BGC in S. albus was targeted (62). A cluster-specific transcriptional eGFP promoter–reporter construct was used as a readout, leading to the identification of etoposide and ivermectin as elicitors and the discovery of fifteen cryptic metabolites.
The reliance on colorimetric or genetic detection in the first applications limited HiTES to pigments or genetically tractable microorganisms. Recently, alternative genetics-independent readouts have been implemented. Specifically, imaging or high-throughput MS has allowed a molecule-first discovery strategy, while biological assays have afforded a bioactivity-first search protocol (Figure 3b). Xu et al. (63) demonstrated that, upon application of HiTES, the induced metabolomes can be rapidly surveyed with IMS to identify new cryptic metabolites. Coupling HiTES to an IMS detection method led to the discovery of nine cryptic NPs, including the keratinicyclins, a new chemotype within the glycopeptide antibiotics containing an unusual 2-oxazolidone moiety (Figure 4). Very recently, Zhang & Seyedsayamdost (64) demonstrated a matrix-assisted laser desorption/ionization–MS-coupled HiTES approach, which enabled the discovery of cinnapeptin, a new, cyclic nonribosomal peptide. There are several advantages to lever-aging MS-based readouts with HiTES. The approach is genetics independent and can be used for species that are difficult to manipulate genetically; it has, in fact, been applied to streptomycetes and rare actinomycetes (63), the latter being exceptionally challenging for genetic manipulation. Moreover, rather than monitor a single BGC with a genetic reporter, MS-based methods allow all elicited NPs to be monitored, as detected by MS. Indeed, the output of the approach, a 3D map that relates the mass-to-charge ratio and abundance of an NP to the presence of hundreds of candidate elicitors, provides an image of the global secondary metabolome of the bacterium under investigation (Figure 3b).
In addition to pigments, genetic reporters, and IMS, high-throughput growth inhibition assays have also been used to assess production of cryptic metabolites. Moon et al. (65) showed that otherwise-undetectable antibiotic activity could be observed in this fashion in three well-studied actinomycetes. Follow-up work with Streptomyces hiroshimensis facilitated the identification of the taylorflavins (Figure 4), cryptic antibiotics that were selective against Gram-negative bacteria and were induced by the hypertension drug atenolol. Other successful applications of bioactivity-coupled HiTES have also been reported (66, 67).
As alluded to above, HiTES has been used to explore the regulatory pathways that link detection of an elicitor to activation of a silent BGC. With ARC2, Craney et al. (56) identified fatty-acid biosynthesis as the target, specifically the enoyl reductase FabI. Both ARC2 and the structurally related molecule triclosan inhibit this enzyme, and S. coelicolor expressing a triclosan-resistant allele of FabI no longer triggered actinorhodin synthesis in response to the antibiotic. Analogously, low-dose trimethoprim was found to act through dihydrofolate reductase in B. thailandensis to enhance secondary metabolism (60, 68). The presence of the antibiotic partially inhibited tetrahydrofolate and consequently Met biosynthesis, leading to the buildup of the immediate precursors homocysteine and homoserine, which acted as effective elicitors. In addition, a complete regulatory circuit has recently been constructed with piperacillin in B. thailandensis (A. Li, B.K. Okada, P.C. Rosen & M.R. Seyedsayamdost, unpublished results).
The vignettes in this section highlight the advantages and challenges of HiTES. It is genetics independent and can be applied to any culturable microbe. Both the products and regulation of silent BGCs can be elucidated, and products from complex BGCs and those stemming from cross talk between BGCs have been identified. Activity-coupled HiTES has uncovered cryptic, bioactive NPs, and the approach is limited only by the bioactivity assay that can be envisioned. A disadvantage, as with any forward chemical-genetics exercise, is that the target and mechanism of action of the elicitor might not be immediately obvious, meaning that additional studies are needed. But, much as RGMS connects unsuspected genes to the activation of silent BGCs, HiTES links unsuspected small molecules to a chosen BGC. These elicitors may have ecological consequences or origins that remain to be investigated.
Reverse Chemical Genetics
Rather than select elicitors from a library, reverse chemical-genetic methods specifically perturb processes that are important for expression of a BGC. Chief among these is modification of either the translational or transcriptional machinery of the host to increase NP biosynthesis, approaches termed ribosome engineering or RNA polymerase (RNAP) engineering, respectively (Figure 5b). Shima et al. (69) were inspired by the observation that streptomycin-resistant Streptomyces lividans, which was obtained by prolonged exposure to the antibiotic, produced the pigment actinorhodin, whereas the untreated parental wild type did not. The mutation was mapped to Lys88 in RpsL (ribosomal protein S12), with Lys88Glu resulting in the phenotype, whereas Lys43Asn, which is used as a streptomycin marker, did not. The mutated ribosome was found to be structurally more stable under conditions of stress, e.g., amino-acid starvation, and to exhibit higher levels of protein synthesis in stationary phase, including synthesis of the actinorhodin pathway–specific transcriptional regulator (70). Ribosome engineering has been extended to other antibiotics, including erythromycin, gentamicin, tetracycline, and lincomycin, and to additional genera, notably Bacillus cereus and Pseudomonas pyrrocinia (71), thereby establishing it as an effective approach for enhancing expression of silent BGCs. Note that ribosome engineering is, in the customary sense, not a reverse chemical-genetic approach, as mutants are typically selected and screened further. We are including it in this section because of the targeted interference with or alteration of a selected physiological pathway using small molecules and because with certain antibiotics (10, 72), inhibition rather than selection of mutated variants resulted in a similar phenotype.
Targeted and engineered mutations have been extended to RNAP, most successfully with rifampicin-resistant alleles. Perhaps the best application of this strategy was in the identification of the novel piperidamycins (Figure 4). In a large bioactivity-guided screen of 1,068 soil actinomycetes, 43% of Streptomyces strains and 6% of nonstreptomycetes with no observable bioactivity were found to produce antibiotics after acquiring resistance to streptomycin, rifampicin, or gentamicin (73). One of these strains, closely related to Streptomyces mauvecolor, was found to produce the cryptic piperidamycins in 7 out of 45 rifampicin-resistant mutants but none of the 65 streptomycin-resistant mutants. The mutated RNAP was proposed to have increased affinity for promoters of select SM BGCs, including genes involved in piperidamycin biosynthesis. The positive effects of ribosome or RNAP mutations can be further enhanced by sequentially accumulating mutations from multiple selection rounds with different antibiotics (74).
The readouts for cryptic metabolite production with ribosome/RNAP engineering have progressed from phenotypic inspection to genetic reporters and bioactivity assays and, most recently, to modern MS-based metabolomics (Figure 2). Goodwin et al. (75) recently pioneered a self-organizing map metabolomics approach to identify a number of NPs that were overproduced in S. coelicolor after acquiring either streptomycin or rifampicin resistance. In another MS-based metabolomic analysis, unsupervised principal component analysis was used to identify several NPs that were overproduced in streptomycin- or rifampicin-resistant Nocardiopsis sp. FU40 mutants relative to their antibiotic-sensitive progenitors (76). This led to the discovery of mutaxanthenes A–C (Figure 4), which were overproduced in roughly half of the rifampicin-resistant mutants, and the ciromicins (75), which were identified from streptomycin-resistant cells. Ribosome/RNAP engineering, therefore, allowed products from at least two cryptic BGCs to be elicited, detected, and prioritized for isolation.
While modifications to the ribosome and RNAP can be effective in increasing NP output, the phenotype depends on the site and nature of the acquired mutation. Therefore, screening of hundreds of mutant strains is typically required. Indeed, reporter-based strategies have been used to quickly identify antibiotic-resistance mutations that result in upregulation of the BGCs of interest (77). Ribosome/RNAP engineering is best suited for NP discovery in a BGC-agnostic fashion, as it is difficult to predict a priori which promoters will be preferred by the mutated RNAP and which messages will be translated efficiently by the altered ribosome. Beyond these limitations, ribosome/RNAP engineering is an elegant, genetics-free approach for enhancing NP biosynthesis in any culturable microorganism.
ENDOGENOUS ACTIVATION: CULTURE MODALITIES
Monocultures
Small changes in cultivation parameters can have large impacts on both primary and secondary metabolism. This is perhaps best exemplified by the extensive optimization of yeast-culture parameters in the beer industry. During yeast fermentation, a temperature change of <2°C can completely alter the flavor profile of beer (78). Similarly, expression of BGCs in many bacteria can be significantly influenced by subtle temperature changes. For example, transcription of the validamycin BGC in Streptomyces hygroscopicus was greatly enhanced by a 2°C increase in the culture temperature (79). In addition to temperature, nutrient availability can also significantly influence NP synthesis, either by altering primary metabolic pathways to change substrate or energy availability or by influencing BGC expression. For instance, sugar availability can directly influence the regulation of BGCs through the well-studied process of carbon catabolite repression, in which pathways that utilize secondary carbon sources are repressed in the presence of a preferred carbon source (80). This phenomenon was observed as early as the 1950s, when penicillin production was shown to increase more than fivefold after switching carbon sources from glucose to xylose (81). In S. lividans, glucose was shown to downregulate AfsR, a global regulator of secondary metabolism (82). The role of carbon sources and the effects of other nutrients (nitrogen, phosphorus, rare earth metals, etc.) on bacterial antibiotic production have been reviewed elsewhere (71, 83).
Taking advantage of these phenomena, nutrient levels and other culture parameters can be systematically varied to activate production of cryptic metabolites in what has become known as the one strain–many compounds (OSMAC) approach (Figure 6a) (84). OSMAC has enhanced production of several cryptic NPs, most recently the terrosamycins, bioactive polyether ionophores from a streptomycete (85). It has been used in diverse bacterial lineages, including proteobacteria, myxobacteria, several marine microorganisms, and other phyla, and good reviews on these topics are available (86, 87). The advantage of OSMAC is certainly its simplicity; there is hardly a simpler approach than growing a bacterium in different media and inspecting for SM production. On the other hand, simple culture variations are not always able to activate a target BGC, as they rely on systematic but arbitrary changes to media composition. For example, NPs from the industrially important model strain Clostridium cellulolyticum remained undetectable in numerous media. NP biosynthesis was ultimately activated through the addition of sterile-filtered aqueous soil extracts taken from the natural habitat of C. cellulolyticum, resulting in the subsequent discovery of the antimicrobial closthioamide (Figure 6) (88). Much as OSMAC may mimic environmental conditions or stresses, the presence of other microorganisms can also lead to enhanced secondary metabolism that may be difficult to recapitulate otherwise. Coculture methods, reviewed below (see the section titled Cocultures), represent an intriguing approach toward modulating culture conditions with environment-type stimuli.
Figure 6.
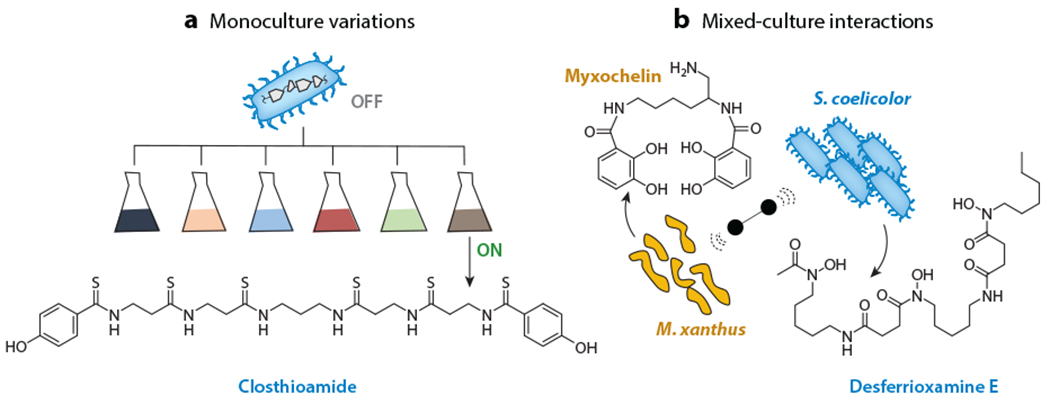
Overview of culture modality approaches. (a) Alterations in microbial culture conditions can significantly impact BGC expression. Monoculture variations screen organisms under several different culture conditions to increase secondary metabolite production. Closthioamide was discovered using this approach. (b) Mixed cultures can activate cryptic BGCs through a multitude of complex interactions between competing and/or cooperating organisms. In this example, production of the siderophores myxochelin by Myxococcus xanthus (shown in gold) and desferrioxamine E by Streptomyces coelicolor (shown in blue) is greatly increased when the two strains are cultured together. Abbreviation: BGC, biosynthetic gene cluster.
Cocultures
Many NPs play an important role in interactions between the producing organism and its host, prey, predators, and/or neighbors. Examples include defensive molecules for self-protection (89, 90) or protection of a mutualistic insect (91), nematode (92), or marine animal (9, 93), as well as antagonistic molecules, which enable parasitism (94) and predation (95). As mediators of such important processes, it is no surprise then, that many BGCs have been elicited by coculture. Analogous to the approaches above, phenotypic and developmental changes, growth inhibition, genetic reporters, and metabolomic approaches have been used to detect cryptic metabolite production, and good reviews are available on these topics (Figure 2) (96, 97). Here, we focus on different types of interactions that can lead to cryptic metabolite production, as well as the advantages and limitations of the approach.
At least four different interaction paradigms are recognized that rely on (a) nutrient changes or depletion, notably those involving iron, (b) signaling molecules, (c) antibiosis through cytotoxic molecules, and (d) direct physical contact. Early studies by Ueda et al. (98) provide a representative example of the first category. A systematic binary interaction assay among 76 streptomycetes showed that 34% of these could act as inducers of antibiotic production (in 13% of receiver strains) or of sporulation (in 9% of receiver strains). A similar assay with wild isolates showed an even higher degree of stimulation with nearly all strains serving as inducers. In follow-up studies, desferrioxamine E produced by Streptomyces griseus was identified as a stimulator of sporulation and development in another streptomycete (Streptomyces tanashiensis) (99). Desferrioxamine and a mixed-ligand siderophore, amychelin, have also been found to mediate the interaction between S. coelicolor and Amycolatopsis sp. AA4 (100, 101). Indeed, iron and siderophores have been found at the center of numerous microbial interactions. When S. coelicolor and the predatory bacterium Myxococcus xanthus are grown nearby on agar, NP biosynthesis in both strains is influenced in part by a reduction in iron availability. M. xanthus initially responds by upregulating genes involved in iron acquisition, including a BGC for the siderophore myxochelin, which sequesters iron (Figure 6b). In response, S. coelicolor upregulates the actinorhodin BGC, which repels M. xanthus (102). While iron deprivation alone is sufficient to activate actinorhodin production in S. coelicolor (103), more actinorhodin is observed in mixed cultures compared to iron-restricted media, indicating that this bacterium may be responding to other interspecies signals from M. xanthus.
Signaling molecules, category b, can be potent elicitors of secondary metabolism. Like many myxobacteria, M. xanthus dedicates a significant portion (>8%) of its genome to the synthesis of NPs, some of which are essential for predation (104), as well as to regulatory networks for cell–cell signaling and small-molecule sensing (105). This bacterium does not produce AHLs, but it can detect and respond to AHL quorum sensing signals from prey bacteria (106). Specifically, its motility and predatory activity are enhanced in response to exogenous AHL signals, while sporulation is delayed. When M. xanthus is cultured adjacent to Escherichia coli on solid agar, myxobacterial NPs such as myxovirescin A and the DKxanthenes are most abundant in the region of predation at the interface between the two species (107). Myxobacterial NPs can also inhibit nearby competitors, especially closely related strains, as mixed cultures of genetically similar M. xanthus strains isolated from the same soil patch are antagonistic (108).
There are many examples in category c (for reviews, see 96, 97), and three representative cases are briefly outlined here. Much like Ochi and colleagues (71), Onaka et al. (109) attempted to induce actinorhodin production from S. lividans, in this case using extracts from 405 actinomycetes. They found a novel thiazole-oxazole-modified microcin, termed goadsporin, that served as an effective elicitor (109). Interestingly, goadsporin was a potent Streptomyces-specific antibiotic. It inhibited growth with good potency, but at subinhibitory concentrations, it elicited actinorhodin production, analogous to the HiTES results described in the section titled Forward Chemical Genetics (58). Similar studies, in which growth inhibition of Bacillus subtilis served as the readout, led to the identification of promomycin, a polyether antibiotic, as a low-dose elicitor and the compound SF-2786 as the induced antibiotic (110). An interesting antagonistic interaction involving a bacteriostatic molecule produced by a fungus and a cryptic fungistatic agent produced by a strep–tomycete, heronapyrrole B, has recently been uncovered in a fungal–streptomycete interaction (111).
Examples of category d, in which physical contact serves as the trigger, have on several occasions been observed in both bacterial–bacterial and fungal–bacterial coculture. Mycolic acid–containing bacteria, which are common coinhabitants of actinomycetes in soil microbiomes, can serve as effective elicitors of cryptic metabolites. For example, mycolic acid–containing bacteria can dramatically increase the pigmentation of S. lividans and other streptomycetes when in direct contact with the SM producer but not when contact is restricted by a dialysis membrane (112). When mycolic-acid synthesis was inhibited, with either antibiotic treatment or gene deletion, pigmentation was greatly reduced. Addition of exogenous mycolic acid did not rescue the phenotype. Further applications of this coculture pairing have led to the discovery of numerous cryptic NPs in both Streptomyces and other actinobacteria, including the cytotoxic butanolides chojalactones (113), the polyenic ciromicins (114), niizalactams (115), and other molecules (Figure 4) (116–118). In the case of orsellinic acid production by Aspergillus nidulans, when cocultured with Streptomyces rapamycinicus, physical contact leading to alterations in fungal chromatin was identified as the key trigger for cryptic NP biogenesis (119, 120).
Coculturing two or more organisms can be a powerful method for activating silent BGCs in any culturable strain. The seminal work of Ueda et al. (98) has recently culminated in the ichip (isolation chip) approach, in which a bacterium, separated by membranes, is cultured in soil to facilitate growth in a native-like environment (121, 122). Coculture can offer clues regarding the chemical ecology and interspecies interactions of the molecules and strains under investigation. Moreover, it is a high-throughput approach, as hundreds of interactions can be screened simultaneously. The disadvantage of this method comes from the stochastic nature of microbial cultures (123). For example, in one study, a mixed culture combination of nine bacteria, none of which produced biologically active compounds in monoculture, would occasionally result in production of the bioactive compound amycomicin, depending on how the community composition developed after inoculation (Figure 4) (124). Even under the most ideal conditions, bacterial growth can vary from one culture to the next, and this is exacerbated under coculture conditions. Moreover, whereas monocultures provide a clear starting point for understanding the nature of BGC activation (iron limitation, carbon source, etc.), mixed cultures require additional follow-up experiments to determine the specific stimuli that ultimately trigger cryptic metabolite biogenesis. Still, as systems approaches are increasingly applied to NP investigations, it is safe to assume that coculture applications will become more prevalent and sophisticated in years to come.
EXOGENOUS ACTIVATION VIA HETEROLOGOUS EXPRESSION
The strategies described so far occur within the native microbial producer. The obvious limitation is that the corresponding microbe must be culturable. It is estimated that the great majority of microorganisms (perhaps ~99%) are not currently culturable under laboratory conditions (125). The only means of accessing their NPs is via cloning and expression of the entire BGC in a heterologous host. This strategy is also well suited for organisms that are culturable but grow poorly or are difficult to culture on a large scale or for silent BGCs from culturable strains that are genetically intractable. The general workflow consists of identifying and mobilizing the target BGC onto a transferable vector, refactoring the regulatory elements, and inserting and expressing the BGC in a suitable exogenous host for isolation of the resulting NP (Figure 7).
Figure 7.
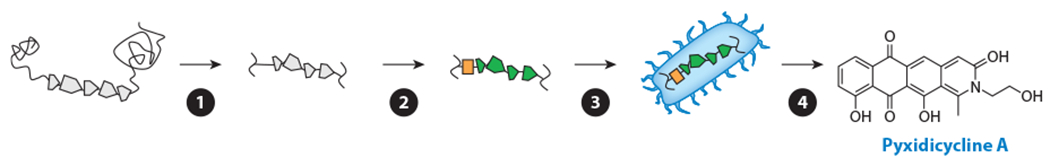
Overview of exogenous activation. Heterologous approaches use a surrogate organism for the production of secondary metabolites from a BGC of interest, usually targeted based on the predicted novelty or utility of the putative natural product. The general steps involved are as follows: ❶ the BGC is identified and assembled into a mobile, transferable vector; ❷ the regulatory elements in the BGC are refactored to suit the surrogate producer; ❸ the mobile, refactored BGC is cloned and expressed in the surrogate host; and ❹ the resultant natural products are identified and characterized. Abbreviation: BGC, biosynthetic gene cluster.
While this is an appealing—and in some cases, the only—strategy for accessing products from silent BGCs, there are some drawbacks. Several programs are available for identifying and annotating NP BGCs. But, determining the boundaries of a BGC is not always straightforward, and it is currently challenging to predict what distal genes are involved or essential for production without a priori knowledge of the structure of the mature NP. A key aspect of heterologous expression is that all of the necessary components that facilitate NP biosynthesis must be made available to the nonnative host. The genotoxin colibactin produced by group B2 E. coli and other pathogenic enterobacteria is representative, as deletion of heat shock protein 90 abolishes colibactin biosynthesis without affecting transcription of the cognate gene cluster (126). Bacterial chaperones encoded outside of a BGC have also been shown to be important for several other NPs (127, 128).
Once identified, there are many methods to mobilize BGCs for bacterial transformation. Relatively small BGCs can be amplified directly from genomic DNA by PCR and joined together by Gibson assembly. Traditionally, cosmid or fosmid libraries were generated in order to piece together medium-sized BGCs. Cosmids are DNA vectors containing λ phage cos regions, which facilitate cyclization after phage-mediated insertion into the heterologous host, and fosmids are similar vectors that also contain genes for the f-factor, which enables bacterial conjugation after transfection. Both of these have been used successfully (129–132) but are limited to BGCs that are 40–50 kb in size. A transformation-associated recombination (TAR) approach, which takes advantage of homologous recombination within yeast, has been used to piece together a small TAR vector with the target BGC (133). This is ultimately used to generate a bacterial artificial chromosome, which can reach several hundred kilobases in length. The TAR approach has facilitated mobilization of silent BGCs and BGCs from unculturable bacteria (133–135), including those coding for taromycin A (136), arixanthomycin (Figure 4) (137), and several other SMs (138, 139). Cas9-assisted targeting of chromosome segments (CATCH) is another modern approach that enables BGCs up to ~150 kb to be captured directly from genomic DNA. Developed by Jiang et al. (140), CATCH uses a Cas9 nuclease to excise a target BGC directly from the genomic DNA, and the entire BGC is then ligated into the cloning vector by Gibson assembly.
During or after mobilization, the native regulators are typically replaced with inducible or constitutively active promoters. Red/ET recombination, using either λ phage proteins Redα and Redβ or Rac phage proteins RecE and RecT, is a homologous recombination approach that can replace (141, 142) or inactivate (143) portions of a BGC. This is particularly important if the heterologous host is not closely related to the native producer. There are also several general promoters that have been identified for heterologous expression in model actinomycete hosts (144, 145). The refactored promoters should ultimately be designed to optimize BGC expression within the selected host. It is also critical to consider relative expression levels of different biosynthetic enzymes, as the right ratio may be essential for successful synthesis of the product (146).
Electroporation and conjugative transfer are commonly used to transport the target DNA into the desired host. The method of choice typically depends on the host and the vector. Even after the BGC is inserted and expressed, the NP may not be observed or captured by the extraction method. Culture conditions can significantly affect NP biosynthesis in a heterologous host, so often multiple growth conditions are tested to identify the best culture parameters. Additionally, the selected surrogate may lack some fundamental metabolic component that would enable production in a different strain, as mentioned with colibactin.
Isolating, mobilizing, refactoring, cloning, and expressing a functional BGC into a surrogate producer is a laborious process, and BGCs targeted by this strategy are often selected based on known or predicted properties of the corresponding NP. For example, a cryptic BGC from the myxobacterium Pyxidicoccus fallax was targeted based on the presence of a pentapeptide repeat protein, a common resistance element for topoisomerase-inhibiting NPs, within the BGC (147). This BGC, under the control of an inserted vanillate promoter, was reconstructed using TAR, and the assembled vector was subsequently inserted and expressed in two phylogenetically related model strains, M. xanthus and Stigmatella aurantiaca. The resulting products, the pyxidicyclines (Figure 7), were indeed found to strongly inhibit E. coli topoisomerase IV and human topoisomerase I. For the malacidins, a different prioritization strategy was used: By analyzing a large cosmid library of metagenomic DNA, Hover et al. (139) identified a clade of putative calcium-dependent antibiotic BGCs that are distantly related to daptomycin and taromycin A, which they predicted would produce novel antibiotics. One of the identified malacidin BGCs was reconstructed using TAR and transformed into S. albus. The products, malacidin A and B, demonstrated clear calcium dependence for antibiosis yet lacked the canonical Asp-X-Asp-Gly motif present in other calcium-dependent antibiotics.
CHOOSING METHODS: A BRIEF GUIDE
Given the diversity of available methods, how does one go about selecting the best approach for a given BGC? First and foremost, whether a cluster is silent needs to be determined by expression analysis (e.g., via reverse transcription quantitative PCR or RNA sequencing) or by HPLCMS, if the product of the BGC or a homologous BGC is known or if an educated guess can be made regarding the structure of the product. A host of methods are available for identifying the products of well-expressed but orphan clusters, which we have not covered here (for reviews, see 148, 149). Once the BGC has been verified as silent or sparingly expressed, the method of choice depends on the complexity of the gene cluster; whether the study is BGC-, metabolite-, or function-centric; and the resources and expertise available to the researchers. In BGC-centric investigations, the complexity of the chosen BGC and its source dictates the method of choice. Small, simple, monooperonic clusters are the best candidates for heterologous approaches. Complex BGCs, however, are best accessed, initially at least, with genetics-free approaches such as OSMAC or HiTES, especially if they are derived from less-studied microbes, which may not be genetically tractable. Medium-sized monooperonic clusters in tractable bacteria can be tackled with both classical genetics and genetics-free strategies. In metabolite-centric projects that prioritize NP structural novelty, methods that use an MS-based readout are preferred. The genetic tractability of the host dictates whether MS-coupled OSMAC, HiTES, coculture, or ribosome engineering is used, or whether classical genetic methods can be employed. Function-centric NP searches are the most flexible, as the bioactivity provides the readout, and any of the activation methods can be applied. Here again, culturability and genetic tractability determine whether exogenous, genetics-dependent endogenous, or genetics-free endogenous methods can be used. Discovery of cryptic metabolites is a two-step process (Figure 3), and routine analytical methods can be employed to troubleshoot each step, where necessary.
SUMMARY AND PERSPECTIVE
Until the turn of the century, microbial NP discovery largely utilized a so-called grind-and-find approach in which cultures were interrogated for readily produced metabolites with the desired bioactivities. In the past 20 years, advances in DNA sequencing and bioinformatics have revealed an abundance of BGCs, many of which are silent or sparingly expressed under standard laboratory conditions and whose products cannot be identified with traditional strategies. Combined approaches from microbial, synthetic, and chemical biology have stepped up to the challenge and delivered methods to mine these BGCs for new cryptic metabolites.
As of today, we have discovered only a tiny fraction of the total arsenal of NPs that has resulted from billions of years of microbial competition, cooperation, and evolution. But, even this small fraction has provided the foundations for modern medicine by identifying privileged drug scaffolds and numerous NP therapeutics. In fact, more than half of the drugs approved by the FDA in the past 40 years can be traced to NPs (2). Moreover, insights into NPs have spurred numerous fields, including NP biosynthesis, total chemical synthesis, chemical ecology, and microbiology. These data underline the impact that successful activation of silent BGCs can have on human health, as well as our understanding of the chemistry and biology of NPs. We have herein described the current strategies for accessing NPs from silent BGCs in bacteria, evaluated their benefits and disadvantages, and provided a useful organizational framework. Moving forward, the continued application of these methods, and the development of new ones, will undoubtedly facilitate the discovery of NPs and NP therapeutics, which are becoming increasingly important due to the spread of multidrug resistance; the appearance of new infectious agents; and a current drug-discovery paradigm that is broadly recognized as slow, expensive, and ineffective. But drug discovery, that is, the medical application of NPs, is not the only arena where we expect cryptic NPs to play a role. Several other aspects will draw significant interest. Foremost among these should be the ecological functions of cryptic NPs and their roles in providing benefits to the host or polymicrobial community, such as in complex soil assemblies or mammalian microbiomes. Beyond function, we still do not have a full understanding of how silent BGCs are regulated, particularly in response to exogenous factors. What kinds of new biochemical transformations remain to be discovered within the multitude of observable orphan BGCs, and, most elusively, what kinds of entirely unique enzymes and novel BGC classes remain unobserved based on the bias and limitations of the available bioinformatic approaches? In addition, NP isolation and structure elucidation are major bottlenecks without major recent progress. High-throughput isolation and structure-elucidation techniques would significantly advance the field. We predict that much future work will focus on these questions and lead to a better understanding of the constituents and functions of the so-called dark matter of microbial metabolomes. The confluence of new methodologies from allied and emerging disciplines clearly points to a renaissance in NP research (150), especially through our ability to exploit, control, and engineer silent BGCs and their products.
BIOSYNTHETIC GENE CLUSTERS: CONSTITUTIVE, SILENT, CRYPTIC, OR ORPHAN?
The terms orphan, cryptic, and silent have been used interchangeably, and sometimes inappropriately, to describe biosynthetic gene clusters (BGCs) identified through genome mining analyses. The term orphan merely indicates that the BGChas not yet been linked to a natural product (NP). Some orphan BGCs may be constitutively expressed and generate readily detectable NPs under standard culture conditions, while the products of others may be difficult to observe. For orphan BGCs that are detected in metagenomic data sets from uncultured bacteria, no assumptions should be made regarding their expression in a hypothetical lab culture.
The products of many BGCs are not readily identifiable under normal culture conditions. These BGCs are termed cryptic, and there could be many underlying reasons, including lack of stability, lack of solubility, lack of an appropriate detection method, or poor expression. This last subset of cryptic BGCs, in which yields of the cognate NP are insufficient due to little or no expression, is known as silent.
HOW MUCH EXPRESSION IS ENOUGH?
How much expression is necessary for detectable levels of a natural product to be synthesized? This depends on the strain, media, and other factors. Orphan biosynthetic gene clusters (BGCs) are assumed to be silent, but this is not always the case. For example, Amos et al. recently observed that staurosporine could be detected at a transcription level of 27.1 reads per kilobase of transcript per million mapped reads (RPKM) but was undetectable at ~12 RPKM (13). When the authors compared the expression of the staurosporine BGC to 48 other BGCs across four Salinispora strains at two different time points, they found that 47% of the BGCs were silent, whereas 53% were expressed at or above the 27.1 RPKM threshold. Interestingly, of the orphan BGCs, 50–69% were transcribed at or above 27.1 RPKM, depending on the strain. Through further exploration, the authors were able to link one of these orphan, yet highly expressed, BGCs to salinipostin.
ACKNOWLEDGMENTS
We are grateful to the National Institutes of Health (grant GM129496 to M.R.S.), the Burroughs Wellcome Fund, the Searle Scholars Program, and the Pew Biomedical Scholars Program for financial support.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Demain AL, Sanchez S. 2009. Microbial drug discovery: 80 years of progress. J. Antibiot 62:5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. 2020. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod 83:770–803 [DOI] [PubMed] [Google Scholar]
- 3.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, et al. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39:W339–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, et al. 2015. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43:9645–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep 26:1362–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugimoto Y, Camacho FR, Wang S, Chankhamjon P, Odabas A, et al. 2019. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome. Science 366:eaax9176. [DOI] [PubMed] [Google Scholar]
- 7.Wilson MR, Zha L, Balskus EP. 2017. Natural product discovery from the human microbiome. J. Biol. Chem 292:8546–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerwick WH, Moore BS. 2012. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol 19:85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita M, Schmidt EW. 2018. Parallel lives of symbionts and hosts: chemical mutualism in marine animals. Nat. Prod. Rep 35:357–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covington BC, Spraggins JM, Ynigez-Gutierrez AE, Hylton ZB, Bachmann BO. 2018. Response of secondary metabolism of hypogean actinobacterial genera to chemical and biological stimuli. Appl. Environ. Microbiol 84:e01125–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derewacz DK, McNees CR, Scalmani G, Covington CL, Shanmugam G, et al. 2014. Structure and stereochemical determination of hypogeamicins from a cave-derived actinomycete. J. Nat. Prod 77:1759–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith TE, Pond CD, Pierce E, Harmer ZP, Kwan J, et al. 2018. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol 14:179–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos GCA, Awakawa T, Tuttle RN, Letzel A-C, Kim MC, et al. 2017. Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. PNAS 114:E11121–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Askenazi M, Driggers EM, Holtzman DA, Norman TC, Iverson S, et al. 2003. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat. Biotechnol 21:150–56 [DOI] [PubMed] [Google Scholar]
- 15.Xiang S-H, Li J, Yin H, Zheng J-T, Yang X, et al. 2009. Application of a double-reporter-guided mutant selection method to improve clavulanic acid production in Streptomyces clavuligerus. Metab. Eng 11:310–18 [DOI] [PubMed] [Google Scholar]
- 16.Guo F, Xiang S, Li L, Wang B, Rajasärkkä J, et al. 2015. Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab. Eng 28:134–42 [DOI] [PubMed] [Google Scholar]
- 17.Xu Z, Wang Y, Chater KF, Ou HY, Xu HH, et al. 2017. Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl. Environ. Microbiol 83:e02889–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed Y, Rebets Y, Tokovenko B, Brötz E, Luzhetskyy A. 2017. Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci. Rep 7:9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JD, Moon K, Miller C, Rose J, Xu F, et al. 2020. Thailandenes, cryptic polyene natural products isolated from Burkholderia thailandensis using phenotype-guided transposon mutagenesis. ACS Chem. Biol 15:1195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao D, Yoshimura A, Wang R, Seyedsayamdost MR. 2020. Reporter-guided transposon mutant selection for activation of silent gene clusters in Burkholderia thailandensis. ChemBioChem 21:1826–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimura A, Covington B, Gallant E, Zhang C, Li A, Seyedsayamdost M. 2020. Unlocking cryptic metabolites using mass spectrometry–guided transposon mutant selection. ACS Chem. Biol 15:2766–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermenau R, Mehl JL, Ishida K, Dose B, Pidot SJ, et al. 2019. Genomics-driven discovery of NO-donating diazeniumdiolate siderophores in diverse plant-associated bacteria. Angew. Chem. Int. Ed 58:13024–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Tian W, Wang S, Yan X, Jia X, et al. 2017. Activation of natural products biosynthetic pathways via a protein modification level regulation. ACS Chem. Biol 12:1732–36 [DOI] [PubMed] [Google Scholar]
- 24.Horinouchi S, Beppu T. 1993. A-factor and streptomycin biosynthesis in Streptomyces griseus. Anton. Leeuw. Int. J. G 64:177–86 [DOI] [PubMed] [Google Scholar]
- 25.Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell. Biol 3:685–95 [DOI] [PubMed] [Google Scholar]
- 26.Maddocks SE, Oyston PCF. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–23 [DOI] [PubMed] [Google Scholar]
- 27.Cai X, Teta R, Kohlhaas C, Crüsemann M, Ueoka R, et al. 2013. Manipulation of regulatory genes reveals complexity and fidelity in hormaomycin biosynthesis. Chem. Biol 20:839–46 [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Wang L, He X, Tian Y, Liu G, Tan H. 2011. SabR enhances nikkomycin production via regulating the transcriptional level of sanG, a pathway-specific regulatory gene in Streptomyces ansochromogenes. BMC Microbiol. 11:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause J, Handayani I, Blin K, Kulik A, Mast Y. 2020. Disclosing the potential of the SARP-type regulator PapR2 for the activation of antibiotic gene clusters in streptomycetes. Front. Microbiol 11:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniel-Ivad M, Hameed N, Tan S, Dhanjal R, Socko D, et al. 2017. An engineered allele of afsQ1 facilitates the discovery and investigation of cryptic natural products. ACS Chem. Biol 12:628–34 [DOI] [PubMed] [Google Scholar]
- 31.Xie C, Deng J-J, Wang H-X. 2015. Identification of AstG1, a LAL family regulator that positively controls ansatrienins production in Streptomyces sp. XZQH13. Curr. Microbiol 70:859–64 [DOI] [PubMed] [Google Scholar]
- 32.Laureti L, Song L, Huang S, Corre C, Leblond P, et al. 2011. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. PNAS 108:6258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanapipatsiri A, Gomez-Escribano JP, Song L, Bibb MJ, Al-Bassam M, et al. 2016. Discovery of unusual biaryl polyketides by activation of a silent Streptomyces venezuelae biosynthetic gene cluster. ChemBioChem 17:2189–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z, Xu Q, Bu Q, Guo Y, Liu S, et al. 2015. Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. ChemBioChem 16:496–502 [DOI] [PubMed] [Google Scholar]
- 35.Biggins JB, Gleber CD, Brady SF. 2011. Acyldepsipeptide HDAC inhibitor production induced in Burkholderia thailandensis. Org. Lett 13:1536–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn SK, Cuthbertson L, Nodwell JR. 2012. Genome context as a predictive tool for identifying regulatory targets of the TetR family transcriptional regulators. PLOS ONE 7:e50562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol. Mol. Biol. Rev 77:440–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidda JD, Song L, Poon V, Al-Bassam M, Lazos O, et al. 2014. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem. Sci 5:86–89 [Google Scholar]
- 39.Zhang Y, Zou Z, Niu G, Tan H. 2013. jadR* and jadR2 act synergistically to repress jadomycin biosynthesis. Sci. China Life Sci 56:584–90 [DOI] [PubMed] [Google Scholar]
- 40.Bunet R, Song L, Mendes MV, Corre C, Hotel L, et al. 2011. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J. Bacteriol 193:1142–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Guo F, Dong SH, Zhao H. 2019. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol 15:111–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, et al. 2008. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higo A, Hara H, Horinouchi S, Ohnishi Y.2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19:259–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenzie NL, Thaker M, Koteva K, Hughes DW, Wright GD, Nodwell JR. 2010. Induction of antimicrobial activities in heterologous streptomycetes using alleles of the Streptomyces coelicolor gene absA1. J. Antibiot 63:177–82 [DOI] [PubMed] [Google Scholar]
- 45.Gehrke EJ, Zhang X, Pimentel-Elardo SM, Johnson AR, Rees CA, et al. 2019. Silencing cryptic specialized metabolism in Streptomyces by the nucleoid-associated protein Lsr2. eLife 8:e47691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao D, Bushin LB, Moon K, Wu Y, Seyedsayamdost MR. 2017. Discovery of scmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. PNAS 114:E2920–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta A, Bedre R, Thapa SS, Sabrin A, Wang G, et al. 2017. Global awakening of cryptic biosynthetic gene clusters in Burkholderia thailandensis. ACS Chem. Biol 12:3012–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Zhang J, Zhuo J, Li Y, Tian Y, Tan H. 2017. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem 292:19708–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nazari B, Kobayashi M, Saito A, Hassaninasab A, Miyashita K, Fujii T. 2013. Chitin-induced gene expression in secondary metabolic pathways of Streptomyces coelicolor A3(2) grown in soil. Appl. Environ. Microbiol 79:707–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao CH, Xu Y, Rigali S, Ye BC. 2015. DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol 99:10215–24 [DOI] [PubMed] [Google Scholar]
- 51.Mukherji R, Zhang S, Chowdhury S, Stallforth P 2020. Chimeric LuxR transcription factors rewire natural product regulation. Angew. Chem. Int. Ed 59:6192–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olano C, García I, González A, Rodriguez M, Rozas D, et al. 2014. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol 7:242–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trottmann F, Franke J, Richter I, Ishida K, Cyrulies M, et al. 2019. Cyclopropanol warhead in malleicyprol confers virulence of human- and animal-pathogenic Burkholderia species. Angew. Chem. Int. Ed 58:14129–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biggins JB, Kang HS, Ternei MA, DeShazer D, Brady SF. 2014. The chemical arsenal of Burkholderia pseudomallei is essential for pathogenicity. J. Am. Chem. Soc 136:9484–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang MM, Wong FT, Wang Y, Luo S, Lim YH, et al. 2017. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol 13:607–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Craney A, Ozimok C, Pimentel-Elardo SM, Capretta A, Nodwell JR. 2012. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol 19:1020–27 [DOI] [PubMed] [Google Scholar]
- 57.Seyedsayamdost MR. 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. PNAS 111:7266–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol 9:445–53 [DOI] [PubMed] [Google Scholar]
- 59.Okada BK, Wu Y, Mao D, Bushin LB, Seyedsayamdost MR. 2016. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem. Biol 11:2124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li A, Mao D, Yoshimura A, Rosen PC, Martin WL, et al. 2020. Multi-omic analyses provide links between low-dose antibiotic treatment and induction of secondary metabolism in Burkholderia thailandensis. mBio 11:e03210–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klaus JR, Deay J, Neuenswander B, Hursh W, Gao Z, et al. 2018. Malleilactone is a Burkholderia pseudomallei virulence factor regulated by antibiotics and quorum sensing. J. Bacteriol 200:e00008–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR. 2017. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc 139:9203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu F, Wu Y, Zhang C, Davis KM, Moon K, et al. 2019. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol 15:161–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C, Seyedsayamdost MR. 2020. Discovery of a cryptic depsipeptide from Streptomyces ghanaensis via MALDI-MS-guided high-throughput elicitor screening. Angew. Chem. Int. Ed 59:23005–9 [DOI] [PubMed] [Google Scholar]
- 65.Moon K, Xu F, Zhang C, Seyedsayamdost MR. 2019. Bioactivity-HiTES unveils cryptic antibiotics encoded in actinomycete bacteria. ACS Chem. Biol 14:767–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moon K, Xu F, Seyedsayamdost MR. 2019. Cebulantin, a cryptic lanthipeptide antibiotic uncovered using bioactivity-coupled HiTES. Angew. Chem. Int. Ed 58:5973–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panthee S, Takahashi S, Hayashi T, Shimizu T, Osada H. 2019. β-carboline biomediators induce reveromycin production in Streptomyces sp. SN-593. Sci. Rep 9:5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Truong TT, Seyedsayamdost M, Greenberg EP, Chandler JR. 2015. A Burkholderia thailandensis acyl-homoserine lactone-independent orphan LuxR homolog that activates production of the cytotoxin malleilactone. J. Bacteriol 197:3456–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol 178:7276–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tamehiro N, Hosaka T, Xu J, Hu H, Otake N, Ochi K. 2003. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl. Environ. Microbiol 69:6412–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ochi K, Hosaka T. 2013. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol 97:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imai Y, Sato S, Tanaka Y, Ochi K, Hosaka T. 2015. Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl. Environ. Microbiol 81:3869–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, et al. 2009. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol 27:462–64 [DOI] [PubMed] [Google Scholar]
- 74.Wang G, Hosaka T, Ochi K. 2008. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl. Environ. Microbiol 74:2834–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodwin CR, Covington BC, Derewacz DK, McNees CR, Wikswo JP, et al. 2015. Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps. Chem. Biol 22:661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO. 2013. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. PNAS 110:2336–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Zhao Y, Liu Q, Huang Y, Hu C, Liao G. 2012. Improvement of A21978C production in Streptomyces roseosporus by reporter-guided rpsL mutation selection. J. Appl. Microbiol 112:1095–101 [DOI] [PubMed] [Google Scholar]
- 78.Webersinke F, Klein H, Flieher M, Urban A, Jäger H, Forster C. 2018. Control of fermentation by products and aroma features of beer produced with Scottish ale yeast by variation of fermentation temperature and wort aeration rate. J. Am. Soc. Brew. Chem 76:147–55 [Google Scholar]
- 79.Liao Y, Wei ZH, Bai L, Deng Z, Zhong JJ. 2009. Effect of fermentation temperature on validamycin A production by Streptomyces hygroscopicus 5008. J. Biotechnol 142:271–74 [DOI] [PubMed] [Google Scholar]
- 80.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol 6:613–24 [DOI] [PubMed] [Google Scholar]
- 81.Soltero FV, Johnson MJ. 1953. The effect of the carbohydrate nutrition on penicillin production by Penicillium chrysogenum Q-176. Appl. Microbiol 1:52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim ES, Hong HJ, Choi CY, Cohen SN. 2001.Modulation of actinorhodin biosynthesis in Streptomyces lividans by glucose repression of afsR2 gene transcription. J. Bacteriol 183:2198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rokem JS, Lantz AE, Nielsen J. 2007. Systems biology of antibiotic production by microorganisms. Nat. Prod. Rep 24:1262–87 [DOI] [PubMed] [Google Scholar]
- 84.Bode HB, Bethe B, Höfs R, Zeeck A. 2002. Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem 3:619–27 [DOI] [PubMed] [Google Scholar]
- 85.Sproule A, Correa H, Decken A, Haltli B, Berrué F, et al. 2019. Terrosamycins A and B, bioactive polyether ionophores from Streptomyces sp. RKND004 from Prince Edward Island sediment. Mar. Drugs 17:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Romano S, Jackson SA, Patry S, Dobson ADW 2018. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 16:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Covington BC, McLean JA, Bachmann BO. 2017. Comparative mass spectrometry–based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep 34:6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lincke T, Behnken S, Ishida K, Roth M, Hertweck C. 2010. Closthioamide: an unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew. Chem. Int. Ed 49:2011–13 [DOI] [PubMed] [Google Scholar]
- 89.Klapper M, Götze S, Barnett R, Willing K, Stallforth P 2016. Bacterial alkaloids prevent amoebal predation. Angew. Chem. Int. Ed 55:8944–47 [DOI] [PubMed] [Google Scholar]
- 90.Arp J, Götze S, Mukherji R, Mattern DJ, García-Altares M, et al. 2018. Synergistic activity of cosecreted natural products from amoebae-associated bacteria. PNAS 115:3758–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beemelmanns C, Guo H, Rischer M, Poulsen M. 2016. Natural products from microbes associated with insects. Beilstein . J. Org. Chem 12:314–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tobias NJ, Wolff H, Djahanschiri B, Grundmann F, Kronenwerth M, et al. 2017. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol 2:1676–85 [DOI] [PubMed] [Google Scholar]
- 93.Tianero MD, Balaich JN, Donia MS. 2019. Localized production of defence chemicals by intracellular symbionts of Haliclona sponges. Nat. Microbiol 4:1149–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seyedsayamdost MR, Wang R, Kolter R, Clardy J. 2014. Hybrid biosynthesis of roseobacticides from algal and bacterial precursor molecules. J. Am. Chem. Soc 136:15150–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xiao Y, Wei X, Ebright R, Wall D. 2011. Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol 193:4626–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Okada BK, Seyedsayamdost MR. 2017. Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev 41:19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pettit RK. 2009. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol 83:19–25 [DOI] [PubMed] [Google Scholar]
- 98.Ueda K, Kawai S, Ogawa H, Kiyama A, Kubota T, et al. 2000. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J. Antibiot 53:979–82 [DOI] [PubMed] [Google Scholar]
- 99.Yamanaka K, Oikawa H, Ogawa HO, Hosono K, Shinmachi F, et al. 2005. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 151:2899–905 [DOI] [PubMed] [Google Scholar]
- 100.Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. 2012. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol. Microbiol 86:628–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seyedsayamdost MR, Traxler MF, Zheng S-L, Kolter R, Clardy J. 2011. Structure and biosynthesis of amychelin, an unusual mixed-ligand siderophore from Amycolatopsis sp. AA4. J. Am. Chem. Soc 133:11434–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee N, Kim W, Chung J, Lee Y, Cho S, et al. 2020. Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME J. 14:1111–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coisne S, Béchet M, Blondeau R. 1999. Actinorhodin production by Streptomyces coelicolor A3(2) in iron-restricted media. Lett. Appl. Microbiol 28:199–202 [DOI] [PubMed] [Google Scholar]
- 104.Xiao Y, Wei X, Ebright R, Wall D. 2011. Antibiotic production by myxobacteria plays a role inpredation. J. Bacteriol 193:4626–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. PNAS 103:15200–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lloyd DG, Whitworth DE. 2017. The myxobacterium Myxococcus xanthus can sense and respond to the quorum signals secreted by potential prey organisms. Front. Microbiol 8:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ellis BM, Fischer CN, Martin LB, Bachmann BO, McLean JA. 2019. Spatiochemically profiling microbial interactions with membrane scaffolded desorption electrospray ionization–ion mobility–imaging mass spectrometry and unsupervised segmentation. Anal. Chem 91:13703–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vos M, Velicer GJ. 2009. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus. Curr. Biol 19:1763–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. 2001. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J. Antibiot 54:1036–44 [DOI] [PubMed] [Google Scholar]
- 110.Amano S-I, Morota T, Kano Y-K, Narita H, Hashidzume T, et al. 2010. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot 63:486–91 [DOI] [PubMed] [Google Scholar]
- 111.Khalil ZG, Cruz-Morales P, Licona-Cassani C, Marcellin E, Capon RJ. 2019. Inter-kingdom beach warfare: Microbial chemical communication activates natural chemical defences. ISME J. 13:147–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Onaka H, Mori Y, Igarashi Y, Furumai T.2011. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol 77:400–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hoshino S, Wakimoto T, Onaka H, Abe I. 2015. Chojalactones A–C, cytotoxic butanolides isolated from Streptomyces sp. cultivated with mycolic acid containing bacterium. Org. Lett 17:1501–4 [DOI] [PubMed] [Google Scholar]
- 114.Derewacz DK, Covington BC, McLean JA, Bachmann BO. 2015. Mapping microbial response metabolomes for induced natural product discovery. ACS Chem. Biol 10:1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoshino S, Okada M, Wakimoto T, Zhang H, Hayashi F, et al. 2015. Niizalactams A–C, multicyclic macrolactams isolated from combined culture of Streptomyces with mycolic acid-containing bacterium. J. Nat. Prod 78:3011–17 [DOI] [PubMed] [Google Scholar]
- 116.Hoshino S, Wong CP, Ozeki M, Zhang H, Hayashi F, et al. 2018. Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium. J. Antibiot 71:653–57 [DOI] [PubMed] [Google Scholar]
- 117.Cueto M, Jensen PR, Kauffman C, Fenical W, Lobkovsky E, Clardy J. 2001. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod 64:1444–46 [DOI] [PubMed] [Google Scholar]
- 118.Oh DC, Jensen PR, Kauffman CA, Fenical W. 2005. Libertellenones A–D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg. Med. Chem 13:5267–73 [DOI] [PubMed] [Google Scholar]
- 119.Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, et al. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. PNAS 106:14558–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nützmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, et al. 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. PNAS 108:14282–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, et al. 2010. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol 76:2445–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, et al. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hao Y-Q, Zhao X-F, Zhang D-Y. 2016. Field experimental evidence that stochastic processes predominate in the initial assembly of bacterial communities. Environ. Microbiol 18:1730–39 [DOI] [PubMed] [Google Scholar]
- 124.Pishchany G, Mevers E, Ndousse-Fetter S, Horvath DJ Jr., Paludo CR, et al. 2018. Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. PNAS 115:10124–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schloss PD, Handelsman J. 2005. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 6:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garcie C, Tronnet S, Garénaux A, McCarthy AJ, Brachmann AO, et al. 2016. The bacterial stress-responsive Hsp90 chaperone (HtpG) is required for the production of the genotoxin colibactin and the siderophore yersiniabactin in Escherichia coli. J. Infect. Dis 214:916–24 [DOI] [PubMed] [Google Scholar]
- 127.Washio K, Lim SP, Roongsawang N, Morikawa M. 2010. Identification and characterization of the genes responsible for the production of the cyclic lipopeptide arthrofactin by Pseudomonas sp. MIS38. Biosci. Biotech. Biochem 74:992–99 [DOI] [PubMed] [Google Scholar]
- 128.Vivien E, Megessier S, Pieretti I, Cociancich S, Frutos R, et al. 2005. Xanthomonas albilineans HtpG is required for biosynthesis of the antibiotic and phytotoxin albicidin. FEMS Microbiol. Lett 251:81–89 [DOI] [PubMed] [Google Scholar]
- 129.Hong ST, Carney JR, Gould SJ. 1997. Cloning and heterologous expression of the entire gene clusters for PD 116740 from Streptomyces strain WP 4669 and tetrangulol and tetrangomycin from Streptomyces rimosus NRRL 3016. J. Bacteriol 179:470–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brady SF, Chao CJ, Handelsman J, Clardy J. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett 3:1981–84 [DOI] [PubMed] [Google Scholar]
- 131.Blodgett JAV, Zhang JK, Metcalf WW 2005.Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob. Agents Chemother 49:230–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eliot AC, Griffin BM, Thomas PM, Johannes TW, Kelleher NL, et al. 2008. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem. Biol 15:765–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bonet B, Teufel R, Crüsemann M, Ziemert N, Moore BS. 2015. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod 78:539–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ross AC, Gulland LE, Dorrestein PC, Moore BS. 2015.Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synth. Biol 4:414–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tang X, Li J, Millán-Aguiñaga N, Zhang JJ, O’Neill EC, et al. 2015. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol 10:2841–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, et al. 2014. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. PNAS 111:1957–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kang H-S, Brady SF. 2014. Arixanthomycins A–C: phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem. Biol 9:1267–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kang H-S, Brady SF. 2014. Mining soil metagenomes to better understand the evolution of natural product structural diversity: pentangular polyphenols as a case study. J. Am. Chem. Soc 136:18111–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hover BM, Kim SH, Katz M, Charlop-Powers Z, Owen JG, et al. 2018. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol 3:415–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang W, Zhao X, Gabrieli T, Lou C, Ebenstein Y, Zhu TF. 2015. Cas9-assisted targeting of chromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun 6:8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tu Q, Herrmann J, Hu S, Raju R, Bian X, et al. 2016. Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via Red/ET recombineering. Sci. Rep 6:21066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Horbal L, Marques F, Nadmid S, Mendes MV, Luzhetskyy A. 2018. Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab. Eng 49:299–315 [DOI] [PubMed] [Google Scholar]
- 143.Bian X, Plaza A, Yan F, Zhang Y, Müller R. 2015. Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo. Biotechnol. Bioeng 112:1343–53. [DOI] [PubMed] [Google Scholar]
- 144.Li S, Wang J, Li X, Yin S, Wang W, Yang K. 2015. Genome-wide identification and evaluation of constitutive promoters in streptomycetes. Microb. Cell. Fact 14:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Luo Y, Zhang L, Barton KW, Zhao H. 2015. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth. Biol 4:1001–10 [DOI] [PubMed] [Google Scholar]
- 146.Smanski MJ, Bhatia S, Zhao D, Park Y, Woodruff LBA, et al. 2014. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol 32:1241–49 [DOI] [PubMed] [Google Scholar]
- 147.Panter F, Krug D, Baumann S, Müller R. 2018. Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria. Chem. Sci 9:4898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Billingsley JM, DeNicola AB, Tang Y. 2016. Technology development for natural product biosynthesis in Saccharomyces cerevisiae. Curr. Opin. Biotechnol 42:74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Krug D, Müller R. 2014. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Product. Rep 31:768–83 [DOI] [PubMed] [Google Scholar]
- 150.Bachmann BO, Van Lanen SG, Baltz RH. 2014. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol 41:175–84 [DOI] [PMC free article] [PubMed] [Google Scholar]


