Figure 49.
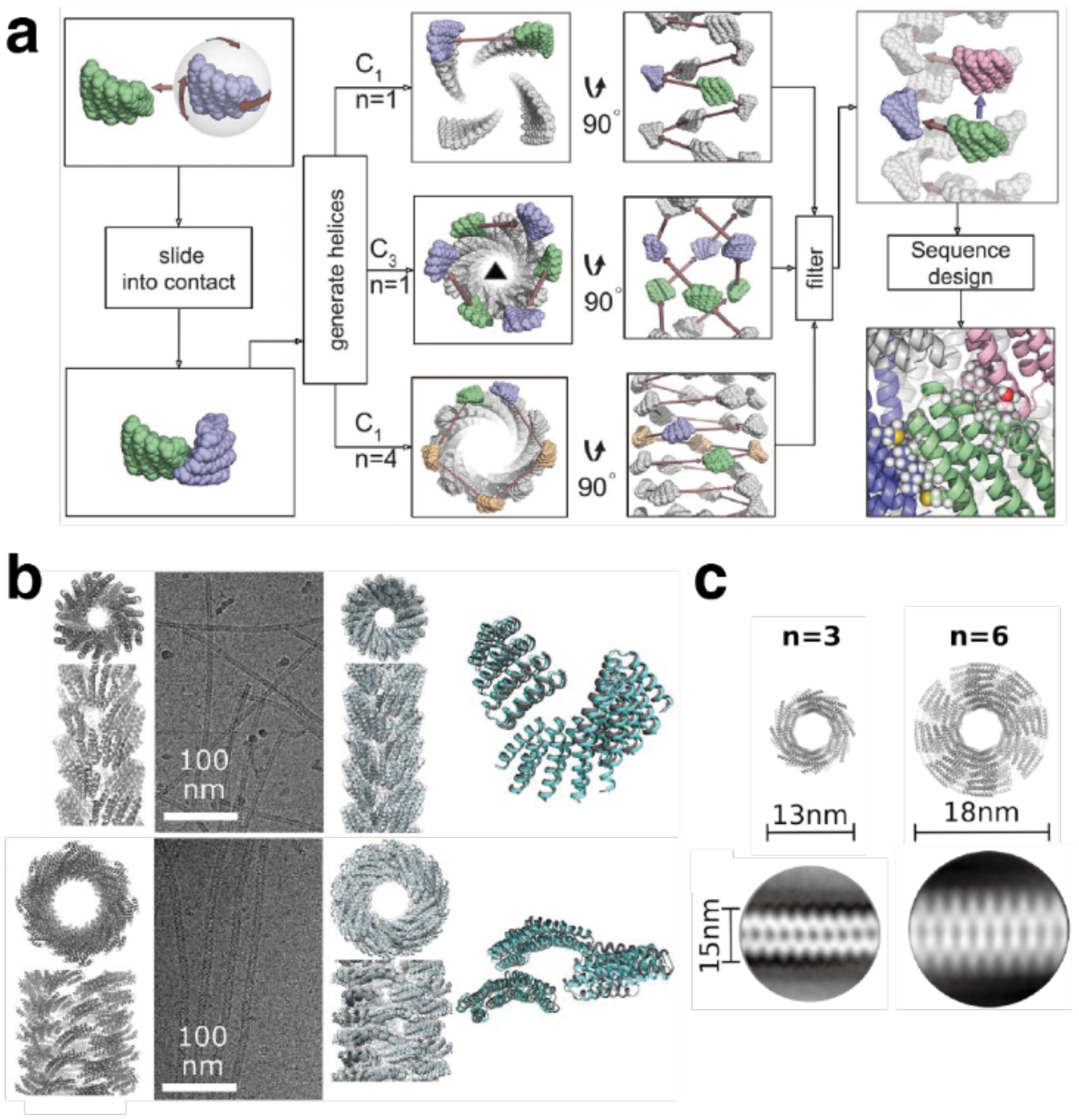
Computationally designed helical protein filaments. a) Computational protocol to design self-assembling protein filaments. A copy of an asymmetric protein monomer is randomly rotated and moved, then slid into contact with the first monomer. The operation is repeated to generate helices. Cyclic symmetry and the ordering of contacting units are screened, and sequence of the monomer is redesigned to optimize the interfaces. b) From left to right: Computationally designed models, cryo-EM micrographs, cryo-EM structures, and overlay of designed model and cryo-EM structure for the C3 symmetric DHF91 (top) and the C1 symmetric DHF79 (bottom) designs. c) Fibers with variable diameter can be generated by changing the number of repeat units within the monomer. Computationally designed models (top) and 2D class average structures (bottom) are shown for two variants of the DHF58 filament. Adapted with permission from Ref.187. Copyright 2018 AAAS.
