ABSTRACT
Nervous system assembly relies on a diversity of cellular processes ranging from dramatic tissue reorganization to local, subcellular changes all driven by precise molecular programs. Combined, these processes culminate in an animal's ability to plan and execute behaviors. Animal behavior can, therefore, serve as a functional readout of nervous system development. Benefitting from an expansive and growing set of molecular and imaging tools paired with an ever-growing number of assays of diverse behaviors, the zebrafish system has emerged as an outstanding platform at the intersection of nervous system assembly, plasticity and behavior. Here, we summarize recent advancements in the field, including how developing neural circuits are refined to shape complex behaviors and plasticity.
KEY WORDS: Behavior, Nervous system, Phenotype, Plasticity, Zebrafish
Summary: This Spotlight summarizes recent work using behavioral analysis methods in embryonic and larval zebrafish that have provided insights into a range of processes crucial for functional nervous system development and refinement.
Introduction
Neuronal proliferation, fate specification and migration, as well as neurite guidance, arborization (branching) and synapse formation, are classical developmental processes underlying the assembly of the nervous system. Each phase of neurodevelopment is orchestrated by diverse molecular-genetic mechanisms that, in turn, coordinate a dizzying array of intra- and intercellular interactions. The ultimate output of these processes is a functional nervous system, capable of detecting and responding to stimuli within the environment, and able to modify those responses as a result of experience. Many of the molecular-genetic regulators of neurodevelopment have been identified through screens in invertebrates, in which the basic processes of neurodevelopment can be observed in vivo, in fixed tissue or even as they occur in real time (Chisholm et al., 2016; Dickson, 2002). However, nervous system development is an ongoing process that does not end with the formation of synapses. Processes including the establishment of mature electrophysiological properties and the strengthening or weakening of synapses through experience also play crucial roles. How can we understand these processes better without directly visualizing them unfolding?
Although the zebrafish is a well-established and powerful system in which to examine directly the classical processes underlying nervous system development (Hutson and Chien, 2002), a large and growing body of literature suggests that examination of zebrafish behavior provides a complementary approach to understanding neurodevelopment. Apart from optical transparency during embryonic and larval stages, there are a variety of features that distinguish the zebrafish as an ideal system for the study of neurodevelopment and behavior. The zebrafish genome is well conserved with respect to humans, with 70% of human genes and 82% of disease-associated genes represented in the zebrafish genome by one or more orthologs (Howe et al., 2013). To evaluate the impact of genes on neurodevelopment and behavior, genetic tools (e.g. CRISPR/Cas9 genome editing) are well developed in the fish (Hwang et al., 2013) and large clutch sizes facilitate high-throughput, rigorous behavioral analysis (Burgess and Granato, 2007; Rihel et al., 2010; Thyme et al., 2019; Wolman et al., 2011). The zebrafish nervous system exhibits significant organizational homology with respect to those of other vertebrates (Kozol et al., 2016), and zebrafish exhibit a wide variety of complex behaviors (reviewed by Fero et al., 2011) with considerable relevance for human disease (Sakai et al., 2018; Vaz et al., 2019). Numerous studies have leveraged the reduced complexity of the larval nervous system by ablating specific neuronal subtypes and identifying specific behavioral consequences, achieving a circuit-level understanding that approaches the single-cell resolution observed in invertebrate systems (Gahtan et al., 2005; Huang et al., 2013; Lacoste et al., 2015; Liu and Fetcho, 1999; McLean et al., 2007; Orger et al., 2008).
Forward genetic screens for uncoordinated (unc) phenotypes in Caenorhabditis elegans have revealed neurodevelopmental regulatory genes ranging from homeobox genes, guidance cues and genes regulating synaptic targeting (Brenner, 1974). Zebrafish genetic screens have been built on this concept (Grunwald and Eisen, 2002); through examination of behavior, screens have highlighted significant conservation of the molecular regulators of neurodevelopment and identified additional vertebrate-specific mechanisms for building a functional nervous system (Granato et al., 1996; Muto et al., 2005; Neuhauss et al., 1999). In addition to providing a platform for unbiased gene discovery, assays of zebrafish behavior provide a test environment to examine hypotheses formed based on developmental studies. For example, studies of nervous system regeneration often use animal behavior as a readout of whether regeneration is functional (Harvey et al., 2019); in the same way, one can ask whether or how a particular developmental perturbation impacts nervous system function. Finally, zebrafish behavior has been successfully leveraged to model human neurodevelopmental disease: genes that are identified through next-generation sequencing in patients can be knocked out in the fish and behavioral consequences can be rigorously examined, often in a high-throughput manner (reviewed by de Abreu et al., 2020; Sakai et al., 2018; Vaz et al., 2019).
In this Spotlight, we highlight recent studies in the embryonic and larval zebrafish that exploit these unique advantages to uncover processes underlying functional nervous system development. First, we summarize how behavioral development over the embryonic and larval period is accomplished through the integration of new circuit modules during nervous system assembly (Pujala and Koyama, 2019). In addition, we highlight studies that reveal how the sensory environment in early life influences circuit development and function (Avitan et al., 2017; Groneberg et al., 2020). Next, we present studies that reveal how behavioral analysis can provide context for neurodevelopment, often using genetic mutants to identify processes that are crucial for function (Asante et al., 2021; Horstick et al., 2020). Finally, we underscore the importance of post-translational modifications and, in particular, the positioning of crucial signaling molecules within the appropriate neuronal subcompartments as a final step in building functional circuits (Carmean et al., 2015; Jain et al., 2018; Lin et al., 2016; Meserve et al., 2021; Nelson et al., 2020).
Behavioral development in the fish is mediated by layering of new circuits onto existing circuitry
Zebrafish begin exhibiting rudimentary behaviors (e.g. spontaneous coiling) on the first day post-fertilization (dpf) (Granato et al., 1996; Saint-Amant and Drapeau, 1998) (Table 1, Fig. 1). These movements are termed ‘spontaneous’ because they occur in the absence of any stimuli, but embryos begin to interact with their sensory environment as they grow. In particular, by 3 dpf, zebrafish respond to touch with characteristic ‘escape’ and ‘struggle’ responses (Liao and Fetcho, 2008; Pujala and Koyama, 2019). Behavioral complexity increases dramatically after 4 dpf (Table 1, Fig. 1) when larvae are able to adapt to their particular surroundings, optimizing their hunting strategies to exploit available prey (Lagogiannis et al., 2020; Oldfield et al., 2020), habituating to repeated stimuli (Best et al., 2008), and adjusting their behavioral thresholds to account for the presence of their siblings (Burgess and Granato, 2008) (Table 1, Fig. 1). We do not yet fully understand how the simple neural circuits that permit early behaviors are modified to accommodate this explosion in behavioral complexity.
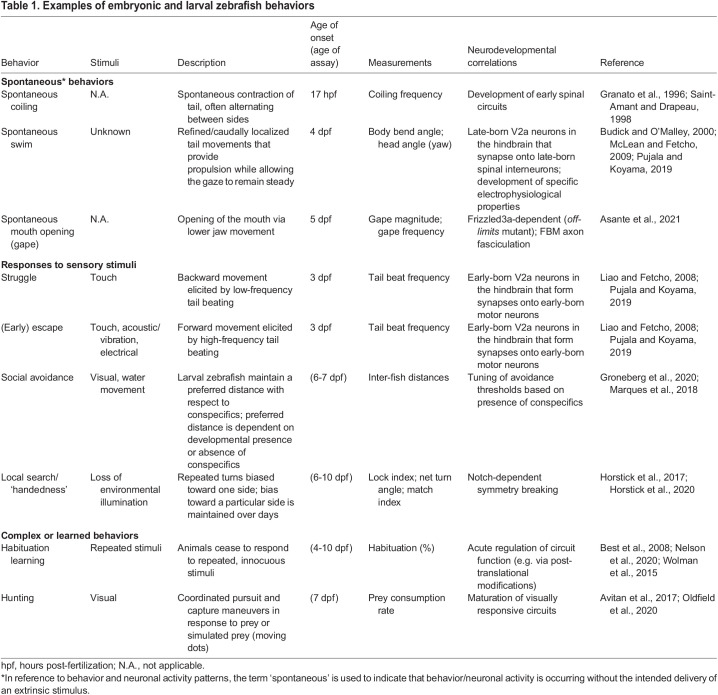
Table 1.
Examples of embryonic and larval zebrafish behaviors
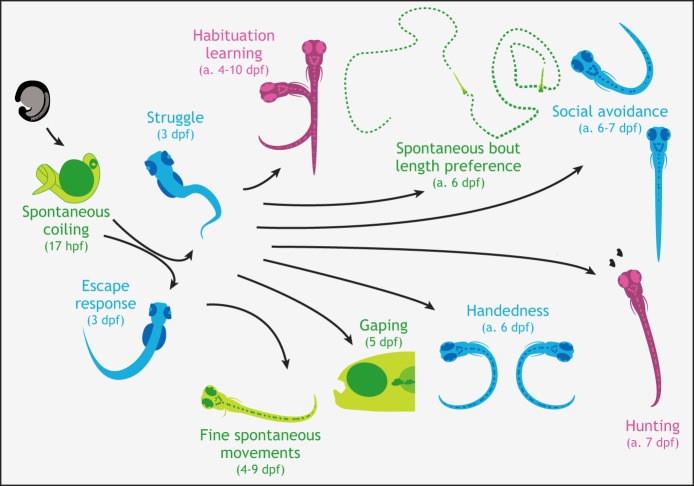
Fig. 1.
Behavioral complexity increases as the nervous system develops. Throughout development, animals perform spontaneous movements (green), which occur without the delivery of an extrinsic stimulus. The first behaviors to emerge are spontaneous coiling movements, which become frequent before the end of the first day post-fertilization. Further development leads to the ability of larvae to perform simple responses to environmental stimuli (blue), including whole-body responses (such as struggles and escapes) to noxious stimuli, which can be evoked by 3 dpf. As larvae develop, behavior becomes more complex and animals gain the ability to modify their responses to stimuli by learning (pink). They additionally gain the ability to respond to a broader range of stimuli (ranging from whole-field illumination changes to the presence of members of their own species) and acquire the ability to more finely tune their spontaneous movements. Recent studies have traced these functional developments back to neurodevelopmental processes. The age of onset (colored text) or age at which a given behavior is assayed (indicated by ‘a.’) is indicated next to each schematized behavior. hpf, hours post-fertilization.
In 2019, Pujala and colleagues set out to understand how zebrafish larvae transition from exclusively performing strong, whole-body movements (e.g. escapes and struggles) (Table 1, Fig. 1) to acquiring the ability to perform weaker and more refined, apparently spontaneous movements (Pujala and Koyama, 2019). They found that sequentially born subsets of V2a neurons in the hindbrain exhibit distinct electrophysiological properties and are selectively active during strong movements (early born) or weak movements (late born). Moreover, they project their axons sequentially to specific targets in the spinal cord, synapsing selectively onto motor neurons mediating strong movements (early born) or interneurons driving weak movements (late born). This work highlights many of the advantages of the zebrafish system, using localized photoconversions, neuronal ablations and in vivo calcium imaging coincident with behavior analysis. In this study, the sequential emergence of two distinct patterns of behavior inspired the discovery of multiple new developmental processes that beg further study. How do axons from sequentially born neuronal populations select different guidance trajectories? How is synaptic partner selection specified in each population? How are distinct electrophysiological properties for each population encoded?
Circuit function feeds back to influence development
Development continues beyond the establishment of functional circuits. Nervous system plasticity permits animals to adapt the form and function of their neural circuits to their surroundings; in the larval zebrafish, early social cues can exert influence on the later functioning of the nervous system. Groneberg and colleagues demonstrated that zebrafish larvae raised in isolation from 3 dpf exhibit enhanced social avoidance and are more easily triggered to perform startle responses upon encountering conspecifics (members of the same species) (Groneberg et al., 2020) (Table 1, Fig. 1). If isolation begins at 0 dpf, even spontaneous movement is affected; isolated fish behave like solitary marathon runners, performing longer and fewer swim bouts compared with their group-raised siblings. As these phenotypes are sensitive to environmental inputs at different developmental time points, they suggest two parallel processes: first, early refinement of locomotor circuitry drives changes in spontaneous movements; and second, later refinements modify sensory thresholds. In this study, careful inspection of the behavior points to the underlying circuitry; enhanced avoidance is partially mediated through visual inputs, but startles are often elicited by conspecifics positioned within the fish's blind spot. Indeed, plasticity arising from non-visual inputs through the lateral line has been confirmed through targeted ablations (Groneberg et al., 2020).
Larval behavior is also sensitive to the presence of prey. Larvae exposed to live prey (e.g. rotifers) become experienced hunters, modifying multiple kinematic parameters of their prey strike behaviors to optimize prey consumption (Lagogiannis et al., 2020). Practice with live prey additionally shapes activity within circuits dedicated to processing visual information (Oldfield et al., 2020). A recent study examined the impact of visual experience on spontaneous neuronal activity in the optic tectum over the developmental period during which zebrafish larvae are ordinarily learning to hunt (4-7 dpf). Avitan and colleagues found that neuronal activity in the tectum increases from 4 dpf until 7 dpf, and then decreases (Avitan et al., 2017), mirroring morphological development in the visual system (early retinal ganglion cell axon arborization, followed by a period of reduced arbor plasticity) (Meyer and Smith, 2006). During this time, spontaneous neuronal activity also becomes more highly correlated between individual neurons in the tectum. Similar to overall activity, the number of assemblies (groups of neurons showing coordinated activity), as well as the number of neurons comprising each assembly, decreases after 5-6 days. Interestingly, inspection of these processes in fish reared in the dark found that measures of neuronal network coordination are reduced relative to their light-reared counterparts. Highlighting the importance of sensory feedback on the development of spontaneous tectal activity, Avitan and colleagues found that dark-reared larvae also have persistent deficits in prey-hunting behavior (Avitan et al., 2017) (Table 1). Together, these studies by Groneberg, Avitan and colleagues highlight the value of studying behavior to understand better how the environment drives neural circuit plasticity during development.
Careful inspection of animal behavior provides context for developmental processes
The establishment of functional connections within the nervous system often involves a complex sequence of events. How are each of these developmental steps relevant for function, and are any of these processes dispensable? Neurons of the facial branchiomotor (FBM) nerve undergo multiple morphological changes that lead to innervation of the jaw and gill muscles in the zebrafish. For example, neurons migrate caudally within the hindbrain then axons are guided rostrally, exit the hindbrain and then arborize over the jaw musculature to form synapses. Later refinement pares back excess arbors and synapses. Interestingly, the functional output of this circuit, jaw movement (Table 1, Fig. 1), does not emerge until long after FBM neurons reach their targets. Asante and colleagues found that the behavioral development closely mirrors later neurodevelopmental processes: jaw gape magnitude increases dramatically between 3 and 5 dpf when FBM axons undergo drastic arborization and synapse formation (Asante et al., 2021). Gape magnitude then recovers to an intermediate level as axon arbors and synapses are pruned, highlighting the functional importance of these processes. Interestingly, the same study found that a genetic mutation in frizzled3a in offlimits mutants, which results in failed caudal migration of FBM neurons (Wada et al., 2006), also results in defasciculation of axons within their target field. As these deficits are associated with defects in jaw gaping and feeding, they suggest that fasciculation is also crucial for establishing a functional circuit.
In a second study, Horstick and colleagues showed that Notch signaling establishes turning side preference in the larval zebrafish (Horstick et al., 2020). Larval zebrafish exhibit stereotyped responses to a sudden transition from whole-field illumination to whole-field darkness. In particular, they perform a localized circling (local search) behavior, in which multiple turns are sequentially performed to one side (Horstick et al., 2017). Although at the population level there was no overall preference for leftward or rightward turning during local searches, individual larvae exhibited turning side preferences (left- or right-‘handedness’) that persisted over several days (Horstick et al., 2020). A small cluster of neurons in the posterior tuberculum, which form a previously unidentified projection within the habenula, are required for the development and/or maintenance of ‘handedness’ in the fish. This work underscores a likely early and stochastic process in the development of nervous system asymmetry. Although the exact nature of this asymmetry is not yet known, further integrative study of the relevant behavior and identified circuit components will undoubtedly reveal the underlying cellular mechanism. This work highlights an intriguing developmental process, the breaking of symmetry, as crucial for circuit function, adding to an existing body of work describing the role of left-right habenular asymmetry in the regulation of anxiety-like behaviors and recovery after exposure to a fearful stimulus (Duboué et al., 2017; Facchin et al., 2015).
Late, local or subtle changes can modulate the emergence of behavior
Synaptic partner selection is often viewed as the final phase in the formation of a functional circuit. Interestingly, the emergence of refined behavior lags temporally after the completion of the ‘classical’ processes of neurodevelopment (e.g. cell migration, axon growth and pathfinding) (Asante et al., 2021; McLean and Fetcho, 2008; Pujala and Koyama, 2019), raising the possibility that behavior finally emerges as a consequence of later, more local and potentially less overt developmental processes. Indeed, the acquisition of mature electrophysiological properties and post-translational modifications of key proteins contribute to behavioral maturation. Nelson and colleagues revealed that the acute activity of the palmitoyltransferase Zdhhc17 (also known as Hip14), is required for the proper function of habituation-regulatory circuits (Nelson et al., 2020). Recently, Meserve and colleagues showed that Dolk, a regulator of glycosylation, is required to position Kv1.1 (encoded by the gene kcna1a), a potassium voltage-gated channel subunit within circuits that regulate locomotor behaviors (Meserve et al., 2021). Furthermore, Jain and colleagues demonstrated that larval responses (selection of a slow versus fast startle) to acoustic stimuli are biased through the regulation of endocytosis of the G protein extracellular calcium-sensing receptor CASR (Jain et al., 2018). Properly positioning these key molecular regulators of behavior represents a final phase in the maturation of neuronal circuits.
Mapping of the touch-insensitive mutant maco to the gene encoding PigK, a member of the GPI anchoring complex, provides another example of the requirement of post-translational modifications for circuit function (Carmean et al., 2015). In this study by Carmean and colleagues, behavior provided a crucial readout, indicating that PigK is required at multiple loci within the touch-sensory circuit. Similarly, a large-scale mutagenesis screen by Lin and colleagues recently identified pinball wizard, a zebrafish mutant with deficits in the acoustic startle response and impaired retinal development (Lin et al., 2016). pinball wizard mapped to wrb (get1), which encodes an endoplasmic reticulum-localized receptor involved in the positioning of tail-anchored proteins within the plasma membrane. In this case, the behavioral phenotypes are attributed to the loss of synaptic expression of Synaptobrevin and Syntaxin 3. In both of these studies by Carmean, Lin and colleagues, unbiased screens for behavioral mutants highlight the importance of post-translational protein processing in achieving a functional nervous system.
Perspectives
Despite recent progress over the past decades, numerous challenges remain regarding the study of behavior and development using the larval zebrafish. In particular, behavioral phenotypes (or lack thereof) identified through the use of morpholinos and CRISPR/Cas9-mediated genome editing must be interpreted with caution (El-Brolosy et al., 2019; Lawson, 2016; Stainier et al., 2017). Going forward, interrogation of the genetic pathways underlying neural circuit development and behavior should aim to eliminate gene promoters or entire gene loci (wherever possible without impinging upon other overlapping genes or gene regulatory elements) to avoid genetic compensation. Furthermore, zebrafish present a challenge to obtaining true cell-type specificity, owing to the limited availability of cell type-specific genetic driver lines. Moreover, considerable transcriptional silencing further limits the utility of the Gal4/UAS system. However, efforts to adapt other transcriptional regulatory strategies, including the Q system (Subedi et al., 2014) and intersectional gene expression strategies to achieve increased specificity (e.g. the use of KillSwitch in Tabor et al., 2018) are underway in fish.
Although there are challenges, we now have an incredible opportunity to understand neurodevelopment through the lens of behavior using the larval zebrafish. Techniques for stimulating, capturing and analyzing zebrafish behavior are becoming cheaper and easier to implement (Joo et al., 2021). Moreover, a wide variety of new tools allow the unprecedented ability to interrogate the development of the nervous system. For example, GRASP (GFP-reconstitution across synaptic partners) has recently been optimized and utilized in fish to demonstrate that inhibitory interneurons in the spinal cord, much like excitatory V2a neurons, target specific motor neuron membrane compartments based on their birth order (Kishore et al., 2020). Viral gene delivery has also been optimized, allowing temporal, genetic and spatial control of gene expression, as well as trans-synaptic circuit tracing (Ma et al., 2020; Satou et al., 2021 preprint). Allowing for a new level of resolution to correlate neuronal position, neurite morphology and even synaptic patterning with function, recent studies have functionally identified neurons based on their activity during a given behavior and subsequently performed electron microscopic reconstruction of the same neurons (Vishwanathan et al., 2017). Additionally, new methods for lineage tracing of unique cell types in the zebrafish brain (McKenna et al., 2016; Raj et al., 2018), together with single-cell RNA sequencing techniques (Raj et al., 2018, 2020), can help us to understand how neuronal fates are specified and how tissues reorganize during development. A handful of new tools allow us to both visualize and manipulate nervous system development and function while assessing the behavioral consequences. Optogenetic (Antinucci et al., 2020; Förster et al., 2017) and whole-brain calcium imaging techniques (reviewed by Vanwalleghem et al., 2018), as well as newly developed genetically encoded voltage indicators (Abdelfattah et al., 2019; Miyazawa et al., 2018), can be combined to probe nervous system function and connectivity in vivo. Light-activated Rac1 allows for the real-time steering of functional axon guidance in vivo in the zebrafish spinal cord (Harris et al., 2020), and neuropeptide-receptor proteins adapted from Hydra permit formation of synthetic synapses between genetically defined cell types (Hawk et al., 2021). Finally, one of the greatest remaining challenges in this field is to use these incredible tools to marry circuit-based and genetic approaches to understand development through the lens of behavior. As we gain more resolution into both circuit architecture and the genes that build those circuits, integrative approaches placing genetic pathways within the relevant circuitry will allow for a holistic understanding of how neuronal circuits are built and how they drive behavior.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the National Institutes of Health (K99NS111736 to J.C.N. and R01NS118921 to M.G.). Deposited in PMC for release after 12 months.
References
- Abdelfattah, A. S., Kawashima, T., Singh, A., Novak, O., Liu, H., Shuai, Y., Huang, Y.-C., Campagnola, L., Seeman, S. C., Yu, J.et al. (2019). Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699-704. 10.1126/science.aav6416 [DOI] [PubMed] [Google Scholar]
- Antinucci, P., Dumitrescu, A., Deleuze, C., Morley, H. J., Leung, K., Hagley, T., Kubo, F., Baier, H., Bianco, I. H. and Wyart, C. (2020). A calibrated optogenetic toolbox of stable zebrafish opsin lines. eLife 9, e54937. 10.7554/eLife.54937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante, E., Hummel, D., Gurung, S., Kassim, Y. M., Al-Shakarji, N., Palaniappan, K., Sittaramane, V. and Chandrasekhar, A. (2021). Defective neuronal positioning correlates with aberrant motor circuit function in zebrafish. Frontiers in Neural Circuits 15, 690475. 10.3389/fncir.2021.690475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitan, L., Pujic, Z., Mölter, J., Van De Poll, M., Sun, B., Teng, H., Amor, R., Scott, E. K. and Goodhill, G. J. (2017). Spontaneous activity in the zebrafish tectum reorganizes over development and is influenced by visual experience. Curr. Biol. 27, 2407-2419.e4. 10.1016/j.cub.2017.06.056 [DOI] [PubMed] [Google Scholar]
- Best, J. D., Berghmans, S., Hunt, J. J. F. G., Clarke, S. C., Fleming, A., Goldsmith, P. and Roach, A. G. (2008). Non-associative learning in larval zebrafish. Neuropsychopharmacology 33, 1206-1215. 10.1038/sj.npp.1301489 [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budick, S. A. and O'Malley, D. M. (2000). Locomotor repertoire of the larval zebrafish: swimming, turning and prey capture. J. Exp. Biol. 203, 2565-2579. 10.1242/jeb.203.17.2565 [DOI] [PubMed] [Google Scholar]
- Burgess, H. A. and Granato, M. (2007). Sensorimotor gating in larval zebrafish. J. Neurosci. 27, 4984-4994. 10.1523/JNEUROSCI.0615-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, H. A. and Granato, M. (2008). The neurogenetic frontier—lessons from misbehaving zebrafish. Brief. Funct. Genomic. Proteomic. 7, 474-482. 10.1093/bfgp/eln039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmean, V., Yonkers, M. A., Tellez, M. B., Willer, J. R., Willer, G. B., Gregg, R. G., Geisler, R., Neuhauss, S. C. and Ribera, A. B. (2015). pigk Mutation underlies macho behavior and affects Rohon-Beard cell excitability. J. Neurophysiol. 114, 1146-1157. 10.1152/jn.00355.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, A. D., Hutter, H., Jin, Y. and Wadsworth, W. G. (2016). The genetics of axon guidance and axon regeneration in caenorhabditis elegans. Genetics 204, 849-882. 10.1534/genetics.115.186262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Abreu, M. S., Genario, R., Giacomini, A. C. V. V., Demin, K. A., Lakstygal, A. M., Amstislavskaya, T. G., Fontana, B. D., Parker, M. O. and Kalueff, A. V. (2020). Zebrafish as a model of neurodevelopmental disorders. Neuroscience 445, 3-11. 10.1016/j.neuroscience.2019.08.034 [DOI] [PubMed] [Google Scholar]
- Dickson, B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959-1964. 10.1126/science.1072165 [DOI] [PubMed] [Google Scholar]
- Duboué, E. R., Hong, E., Eldred, K. C. and Halpern, M. E. (2017). Left habenular activity attenuates fear responses in larval zebrafish. Curr. Biol. 27, 2154-2162.e3. 10.1016/j.cub.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy, M. A., Kontarakis, Z., Rossi, A., Kuenne, C., Günther, S., Fukuda, N., Kikhi, K., Boezio, G. L. M., Takacs, C. M., Lai, S.-L.et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193-197. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchin, L., Duboue, E. R. and Halpern, M. E. (2015). Disruption of epithalamic left-right asymmetry increases anxiety in Zebrafish. J. Neurosci. 35, 15847-15859. 10.1523/JNEUROSCI.2593-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fero, K., Yokogawa, T. and Burgess, H. A. (2011). The Behavioral Repertoire of Larval Zebrafish. In Zebrafish Models in Neurobehavioral Research (ed. Kalueff A. V. and Cachat J. M.), pp. 249-291. Totowa, NJ: Humana Press. [Google Scholar]
- Förster, D., Dal Maschio, M., Laurell, E. and Baier, H. (2017). An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nat. Commun. 8, 116. 10.1038/s41467-017-00160-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahtan, E., Tanger, P. and Baier, H. (2005). Visual prey capture in larval zebrafish is controlled by identified reticulospinal neurons downstream of the tectum. J. Neurosci. 25, 9294-9303. 10.1523/JNEUROSCI.2678-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato, M., Furutani-Seiki, M., Haffter, P., Hammerschmidt, M., Heisenberg, C.-P., Jiang, Y.-J., Kane, D. A., Kelsh, R. N., Mullins, M. C., Odenthal, J.et al. (1996). Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 123, 399-413. 10.1242/dev.123.1.399 [DOI] [PubMed] [Google Scholar]
- Groneberg, A. H., Marques, J. C., Martins, A. L., Diez del Corral, R., de Polavieja, G. G. and Orger, M. B. (2020). Early-life social experience shapes social avoidance reactions in larval zebrafish. Curr. Biol. 30, 4009-4021.e4. 10.1016/j.cub.2020.07.088 [DOI] [PubMed] [Google Scholar]
- Grunwald, D. J. and Eisen, J. S. (2002). Headwaters of the zebrafish — emergence of a new model vertebrate. Nat. Rev. Genet. 3, 717-724. 10.1038/nrg892 [DOI] [PubMed] [Google Scholar]
- Harris, J. M., Wang, A. Y.-D., Boulanger-Weill, J., Santoriello, C., Foianini, S., Lichtman, J. W., Zon, L. I. and Arlotta, P. (2020). Long-range optogenetic control of axon guidance overcomes developmental boundaries and defects. Dev. Cell 53, 577-588.e7. 10.1016/j.devcel.2020.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, B. M., Baxter, M. and Granato, M. (2019). Optic nerve regeneration in larval zebrafish exhibits spontaneous capacity for retinotopic but not tectum specific axon targeting. PLoS ONE 14, e0218667. 10.1371/journal.pone.0218667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk, J. D., Wisdom, E. M., Sengupta, T., Kashlan, Z. D. and Colón-Ramos, D. A. (2021). A genetically encoded tool for reconstituting synthetic modulatory neurotransmission and reconnect neural circuits in vivo. Nat. Commun. 12, 4795. 10.1038/s41467-021-24690-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstick, E. J., Bayleyen, Y., Sinclair, J. L. and Burgess, H. A. (2017). Search strategy is regulated by somatostatin signaling and deep brain photoreceptors in zebrafish. BMC Biol. 15, 4. 10.1186/s12915-016-0346-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstick, E. J., Bayleyen, Y. and Burgess, H. A. (2020). Molecular and cellular determinants of motor asymmetry in zebrafish. Nat. Commun. 11, 1170. 10.1038/s41467-020-14965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., Collins, J. E., Humphray, S., McLaren, K., Matthews, L.et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498-503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K.-H., Ahrens, M. B., Dunn, T. W. and Engert, F. (2013). Spinal projection neurons control turning behaviors in zebrafish. Curr. Biol. 23, 1566-1573. 10.1016/j.cub.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson, L. D. and Chien, C.-B. (2002). Wiring the zebrafish: axon guidance and synaptogenesis. Curr. Opin. Neurobiol. 12, 87-92. 10.1016/S0959-4388(02)00294-5 [DOI] [PubMed] [Google Scholar]
- Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., Peterson, R. T., Yeh, J.-R. J. and Joung, J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R. A., Wolman, M. A., Marsden, K. C., Nelson, J. C., Shoenhard, H., Echeverry, F. A., Szi, C., Bell, H., Skinner, J., Cobbs, E. N.et al. (2018). A forward genetic screen in zebrafish identifies the G-protein-coupled receptor CaSR as a modulator of sensorimotor decision making. Curr. Biol. 28, 1357-1369.e5. 10.1016/j.cub.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, W., Vivian, M. D., Graham, B. J., Soucy, E. R. and Thyme, S. B. (2021). A customizable low-cost system for massively parallel zebrafish behavioral phenotyping. Front. Behav. Neurosci. 14, 258. 10.3389/fnbeh.2020.606900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore, S., Cadoff, E. B., Agha, M. A. and McLean, D. L. (2020). Orderly compartmental mapping of premotor inhibition in the developing zebrafish spinal cord. Science 370, 431-436. 10.1126/science.abb4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozol, R. A., Abrams, A. J., James, D. M., Buglo, E., Yan, Q. and Dallman, J. E. (2016). Function over form: modeling groups of inherited neurological conditions in zebrafish. Front. Mol. Neurosci. 9, 55. 10.3389/fnmol.2016.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste, A. M. B., Schoppik, D., Robson, D. N., Haesemeyer, M., Portugues, R., Li, J. M., Randlett, O., Wee, C. L., Engert, F. and Schier, A. F. (2015). A convergent and essential interneuron pathway for mauthner-cell-mediated escapes. Curr. Biol. 25, 1526-1534. 10.1016/j.cub.2015.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagogiannis, K., Diana, G. and Meyer, M. P. (2020). Learning steers the ontogeny of an efficient hunting sequence in zebrafish larvae. eLife 9, e55119. 10.7554/eLife.55119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, N. D. (2016). Reverse genetics in zebrafish: mutants, morphants, and moving forward. Trends Cell Biol. 26, 77-79. 10.1016/j.tcb.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Liao, J. C. and Fetcho, J. R. (2008). Shared versus specialized glycinergic spinal interneurons in axial motor circuits of larval zebrafish. J. Neurosci. 28, 12982-12992. 10.1523/JNEUROSCI.3330-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.-Y., Vollrath, M. A., Mangosing, S., Shen, J., Cardenas, E. and Corey, D. P. (2016). The zebrafish pinball wizard gene encodes WRB, a tail–anchored–protein receptor essential for inner–ear hair cells and retinal photoreceptors. J. Physiol. 594, 895-914. 10.1113/JP271437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K. S. and Fetcho, J. R. (1999). Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron 23, 325-335. 10.1016/S0896-6273(00)80783-7 [DOI] [PubMed] [Google Scholar]
- Ma, M., Kler, S. and Pan, Y. A. (2020). Structural neural connectivity analysis in zebrafish with restricted anterograde transneuronal viral labeling and quantitative brain mapping. Front. Neural Circuits 13, 85. 10.3389/fncir.2019.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, J. C., Lackner, S., Félix, R. and Orger, M. B. (2018). Structure of the zebrafish locomotor repertoire revealed with unsupervised behavioral clustering. Curr. Biol. 28, 181-195.e5. 10.1016/j.cub.2017.12.002 [DOI] [PubMed] [Google Scholar]
- McKenna, A., Findlay, G. M., Gagnon, J. A., Horwitz, M. S., Schier, A. F. and Shendure, J. (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, D. L. and Fetcho, J. R. (2008). Using imaging and genetics in zebrafish to study developing spinal circuits in vivo. Dev. Neurobiol. 68, 817-834. 10.1002/dneu.20617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, D. L. and Fetcho, J. R. (2009). Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J. Neurosci. 29, 13566-13577. 10.1523/JNEUROSCI.3277-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, D. L., Fan, J., Higashijima, S., Hale, M. E. and Fetcho, J. R. (2007). A topographic map of recruitment in spinal cord. Nature 446, 71-75. 10.1038/nature05588 [DOI] [PubMed] [Google Scholar]
- Meserve, J. H., Nelson, J. C., Marsden, K. C., Hsu, J., Echeverry, F. A., Jain, R. A., Wolman, M. A., Pereda, A. E. and Granato, M. (2021). A forward genetic screen identifies Dolk as a regulator of startle magnitude through the potassium channel subunit Kv1.1. PLoS Genet. 17, e1008943. 10.1371/journal.pgen.1008943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M. P. and Smith, S. J. (2006). Evidence from In Vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. J. Neurosci. 26, 3604-3614. 10.1523/JNEUROSCI.0223-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa, H., Okumura, K., Hiyoshi, K., Maruyama, K., Kakinuma, H., Amo, R., Okamoto, H., Yamasu, K. and Tsuda, S. (2018). Optical interrogation of neuronal circuitry in zebrafish using genetically encoded voltage indicators. Sci. Rep. 8, 6048. 10.1038/s41598-018-23906-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, A., Orger, M. B., Wehman, A. M., Smear, M. C., Kay, J. N., Page-McCaw, P. S., Gahtan, E., Xiao, T., Nevin, L. M., Gosse, N. J.et al. (2005). Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 1, e66. 10.1371/journal.pgen.0010066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J. C., Witze, E., Ma, Z., Ciocco, F., Frerotte, A., Randlett, O., Foskett, J. K. and Granato, M. (2020). Acute regulation of habituation learning via posttranslational palmitoylation. Curr. Biol. 30, 2729-2738.e4. 10.1016/j.cub.2020.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauss, S. C., Biehlmaier, O., Seeliger, M. W., Das, T., Kohler, K., Harris, W. A. and Baier, H. (1999). Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J. Neurosci. 19, 8603-8615. 10.1523/JNEUROSCI.19-19-08603.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield, C. S., Grossrubatscher, I., Chávez, M., Hoagland, A., Huth, A. R., Carroll, E. C., Prendergast, A., Qu, T., Gallant, J. L., Wyart, C.et al. (2020). Experience, circuit dynamics, and forebrain recruitment in larval zebrafish prey capture. eLife 9, e56619. 10.7554/eLife.56619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orger, M. B., Kampff, A. R., Severi, K. E., Bollmann, J. H. and Engert, F. (2008). Control of visually guided behavior by distinct populations of spinal projection neurons. Nat. Neurosci. 11, 327-333. 10.1038/nn2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujala, A. and Koyama, M. (2019). Chronology-based architecture of descending circuits that underlie the development of locomotor repertoire after birth. eLife 8, e42135. 10.7554/eLife.42135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, B., Wagner, D. E., McKenna, A., Pandey, S., Klein, A. M., Shendure, J., Gagnon, J. A. and Schier, A. F. (2018). Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442-450. 10.1038/nbt.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, B., Farrell, J. A., Liu, J., El Kholtei, J., Carte, A. N., Navajas Acedo, J., Du, L. Y., McKenna, A., Relić, Đ., Leslie, J. M.et al. (2020). Emergence of neuronal diversity during vertebrate brain development. Neuron 108, 1058-1074.e6. 10.1016/j.neuron.2020.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel, J., Prober, D. A., Arvanites, A., Lam, K., Zimmerman, S., Jang, S., Haggarty, S. J., Kokel, D., Rubin, L. L., Peterson, R. T.et al. (2010). Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348-351. 10.1126/science.1183090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Amant, L. and Drapeau, P. (1998). Time course of the development of motor behaviors in the zebrafish embryo. J. Neurobiol. 37, 622-632. [DOI] [PubMed] [Google Scholar]
- Sakai, C., Ijaz, S. and Hoffman, E. J. (2018). Zebrafish models of neurodevelopmental disorders: past, present, and future. Front. Mol. Neurosci. 11, 294. 10.3389/fnmol.2018.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou, C., Neve, R. L., Oyibo, H. K., Bouldoires, E. A., Mori, T., Higashijima, S., Keller, G. B. and Friedrich, R. W. (2021). A viral toolbox for conditional and transneuronal gene expression in zebrafish. bioRxiv 10.1101/2021.03.25.436574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier, D. Y. R., Raz, E., Lawson, N. D., Ekker, S. C., Burdine, R. D., Eisen, J. S., Ingham, P. W., Schulte-Merker, S., Yelon, D., Weinstein, B. M.et al. (2017). Guidelines for morpholino use in zebrafish. PLoS Genet. 13, e1007000. 10.1371/journal.pgen.1007000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi, A., Macurak, M., Gee, S. T., Monge, E., Goll, M. G., Potter, C. J., Parsons, M. J. and Halpern, M. E. (2014). Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods 66, 433-440. 10.1016/j.ymeth.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor, K. M., Smith, T. S., Brown, M., Bergeron, S. A., Briggman, K. L. and Burgess, H. A. (2018). Presynaptic inhibition selectively gates auditory transmission to the brainstem startle circuit. Curr. Biol. 28, 2527-2535.e8. 10.1016/j.cub.2018.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme, S. B., Pieper, L. M., Li, E. H., Pandey, S., Wang, Y., Morris, N. S., Sha, C., Choi, J. W., Herrera, K. J., Soucy, E. R.et al. (2019). Phenotypic landscape of schizophrenia-associated genes defines candidates and their shared functions. Cell 177, 478-491.e20. 10.1016/j.cell.2019.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwalleghem, G. C., Ahrens, M. B. and Scott, E. K. (2018). Integrative whole-brain neuroscience in larval zebrafish. Curr. Opin. Neurobiol. 50, 136-145. 10.1016/j.conb.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Vaz, R., Hofmeister, W. and Lindstrand, A. (2019). Zebrafish models of neurodevelopmental disorders: limitations and benefits of current tools and techniques. Int. J. Mol. Sci. 20, 1296. 10.3390/ijms20061296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanathan, A., Daie, K., Ramirez, A. D., Lichtman, J. W., Aksay, E. R. F. and Seung, H. S. (2017). Electron microscopic reconstruction of functionally identified cells in a neural integrator. Curr. Biol. 27, 2137-2147.e3. 10.1016/j.cub.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, H., Tanaka, H., Nakayama, S., Iwasaki, M. and Okamoto, H. (2006). Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development 133, 4749-4759. 10.1242/dev.02665 [DOI] [PubMed] [Google Scholar]
- Wolman, M. A., Jain, R. A., Liss, L. and Granato, M. (2011). Chemical modulation of memory formation in larval zebrafish. Proc. Natl. Acad. Sci. USA 108, 15468-15473. 10.1073/pnas.1107156108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman, M. A., Jain, R. A., Marsden, K. C., Bell, H., Skinner, J., Hayer, K. E., Hogenesch, J. B. and Granato, M. (2015). A genome-wide screen identifies PAPP-AA-mediated IGFR signaling as a novel regulator of habituation learning. Neuron 85, 1200-1211. 10.1016/j.neuron.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]