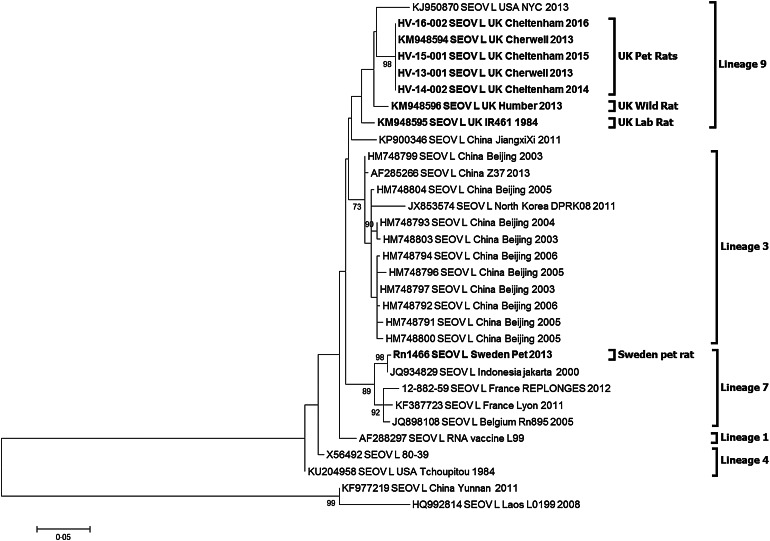
Fig. 1.
Maximum likelihood phylogenetic tree for SEOV partial L segment (333nt) sequences (n = 31) using model Tamura three-parameter model with γ distribution in the MEGA6 package of software with bootstrap of 10 000 [28, 29]. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The scale bar indicates amino acid substitutions per site. Only bootstrap support of >70% are shown. The phylogenetic positions of the UK pet rats are shown in relation to representative Seoul virus strains. Genbank accession numbers are shown next to taxa names.