SUMMARY
Passive surveillance for lyssaviruses in UK bats has been ongoing since 1987 and has identified 13 cases of EBLV-2 from a single species; Myotis daubentonii. No other lyssavirus species has been detected. Between 2005 and 2015, 10 656 bats were submitted, representing 18 species, creating a spatially and temporally uneven sample of British bat fauna. Uniquely, three UK cases originate from a roost at Stokesay Castle in Shropshire, England, where daily checks for grounded and dead bats are undertaken and bat carcasses have been submitted for testing since 2007. Twenty per cent of Daubenton's bats submitted from Stokesay Castle since surveillance began, have tested positive for EBLV-2. Phylogenetic analysis reveals geographical clustering of UK viruses. Isolates from Stokesay Castle are more closely related to one another than to viruses from other regions. Daubenton's bats from Stokesay Castle represent a unique opportunity to study a natural population that appears to maintain EBLV-2 infection and may represent endemic infection at this site. Although the risk to public health from EBLV-2 is low, consequences of infection are severe and effective communication on the need for prompt post-exposure prophylaxis for anyone that has been bitten by a bat is essential.
Key words: Emerging infections, Lyssavirus, rabies (animal), bat, surveillance, Zoonoses
INTRODUCTION
Rabies is caused following exposure and infection by RNA-viruses belonging to the genus Lyssavirus within the Rhabdoviridae family of the Order Mononegavirales. Rabies lyssavirus (RABV) is the type species and is responsible for the majority of human rabies cases. The International Committee on Taxonomy of Viruses delineates the Lyssavirus genus into 14 virus species [1], with two putative species; Lleida bat lyssavirus and Gannoruwa bat lyssavirus awaiting classification [2,3]. Whilst rabies cases caused by lyssavirus species other than RABV are rare, the inability of the most frequently used diagnostic assays to differentiate between lyssavirus species, precludes a meaningful assessment of the human health impact of each of the lyssaviruses [4].
Genetic relationships between lyssaviruses have been well characterized and species belong to one of three phylogroups, based on their genetic relatedness [5]. Within species, lyssavirus strains often cluster geographically, which is unsurprising given the mode of virus transmission; direct contact among host reservoirs. Despite occasional occurrences of spillover into non-reservoir hosts, lyssaviruses circulate within specific host species, and therefore are also restricted by the range of the host.
Viruses from six lyssavirus species (RABV, EBLV-1, EBLV-2, BBLV, WCBV and LLEBV) have been detected in Europe, of which only EBLV-2 has been detected in the UK. Surveillance for EBLV-2, as part of a wider surveillance for all lyssaviruses, is undertaken in a number of countries across Europe, including Belgium, Denmark, Finland, France, Germany, Italy, The Netherlands, Poland, Spain, Sweden, Switzerland and the UK [6,7]. The methodology and coverage of surveillance varies between countries and it is likely that EBLV-2 cases are underestimated because of this inconsistency, low sensitivity of surveillance and lack of reporting. EBLV-2 has been found in Daubenton's bats (Myotis daubentonii) in the UK, Germany, Finland and Switzerland, and in Pond bats (Myotis dasycneme) in The Netherlands [8]. The geographical range of M. dasycneme does not encompass the UK.
There have been two reported lethal EBLV-2 infections in humans [9,10] but no reported cross species transmission events of EBLV-2 from bat species to terrestrial carnivores, as have been described for RABV [11,12] and EBLV-1 [13]. Although the risk to the general public of contracting rabies from an EBLV-2 infected bat is low, the potential for infection remains and consequences are severe. The risk increases, however, for people who have contact with bats for occupational (e.g. veterinarians, bat workers, bat researchers and taxidermists), or recreational purposes (e.g. fly fishermen, bat group members and those interested in bat care and rehabilitation) and are unvaccinated. For this reason it is recommended that bats should only be handled while wearing appropriate bite-proof gloves by handlers vaccinated against rabies; with a known neutralizing antibody titre.
Daubenton's bats are considered relatively common and widespread across the UK [14]. The national population appears to be relatively stable since 1999 [15], which is corroborated by surveys of hibernation sites, however, it is unclear what proportion has been sampled. An extrapolated population size estimate of 150 000 was produced in 1995 [14], and more recently, systematic quantitative ensemble modelling has suggested that the UK Daubenton's bat population is within the range of 39 000–245 000 [16]. No plausible estimate of the mean or median of the population size could be made for this species; in part because of their dependence on waterways which are poorly represented in habitat maps and because of their dependence on trees for roosting.
Daubenton's bats are a medium-sized (body weight approximately 8 g), insectivorous species that are usually associated with roosting and foraging close to water. During the summer they aggregate in maternity roosts, often in trees or bridges and are not generally considered to be a synanthropic species, although they may occasionally also roost within buildings [6]. Daubenton's bats may share roosts with other UK bat species. In the winter, Daubenton's hibernate in underground sites such as caves or mines [15]. Active surveillance studies undertaken in the UK between 2003 and 2012 tested targeted populations of Daubenton's bats, Natterer's bats (Myotis nattereri) and serotines (Eptesicus serotinus) for antibodies against EBLV-2 and EBLV-1, respectively [17–19]. The presence of specific antibodies indicates a previous exposure to virus and does not indicate active clinical or sub-clinical infection at the time of sampling. Seroprevalence in Daubenton's bats at four a-priori sites (EBLV-2 case identified by passive surveillance at that location in the past) sampled was estimated between 2% and 11% [95% confidence interval (CI); mean 5·8%]. In contrast, seroprevalence in Daubenton's bats sampled at random locations (no previous history of EBLV-2, n = 25) was estimated to be substantially lower [on average 2·2% (95% CI, 1·0–4·1%)] [19], suggesting that exposure of Daubenton's bats to EBLV-2 is higher in areas in which bats with clinical rabies have previously been detected. During the same study, oropharyngeal swabs were taken and tested for the presence of viral RNA or infectious virus in saliva, to detect active EBLV infection and to inform assessment of risk to humans. No viral RNA or live virus was detected in any of the swabs in either study [18,19].
Passive surveillance for EBLVs, defined in this instance as the monitoring of infection in dead bats that relies on the submission of samples from the wider public, has been undertaken in the UK since 1987 [20]. The scheme relies on ad hoc submissions of bat carcasses by the general public, veterinarians, the Bat Conservation Trust, associated local bat groups and other non-governmental organizations (NGOs). A previous study described data from the passive bat surveillance scheme between 1987 and 2004 [20], here an update is provided for the subsequent period to 2015. Furthermore, the utility of this surveillance is assessed, with a focus on the first site at which multiple detections of EBLV-2 have been made.
METHODS
Ethics statement exemption
No ethics statement is required for this work on the basis the bats submitted for this work are either found dead or humanely euthanized by a veterinarian as they have been found dying or with severe injuries.
Lyssavirus testing
Bat carcasses were submitted to APHA from across the UK between 2005 and 2015. Key details of the sample reception process including host species identification and essential descriptive data are provided by the submitter using a standardized submission form. More details regarding the submission information provided can be accessed in Supplementary material S1. Only dead bats are accepted for the purpose of passive surveillance, not sick or dying bats. Species identification is carried out by a trained member of staff, using a key based on morphological features [See: The Field Studies Council website at http://www.field-studies-council.org/publications/pubs/bats-identification-chart.aspx (Accessed 30th May 2017)]. A brain sample was obtained from each bat carcass by dissection, performed by a trained member of staff under containment level 3 conditions.
The gold standard fluorescent antibody test (FAT) for lyssavirus diagnosis was carried out as described previously [21]. The in vitro Rabies Tissue Culture Inoculation Test (RTCIT) was used to confirm the presence or absence of live virus as previously described [22].
A semi-quantitative, real-time, TaqMan® reverse-transcriptase polymerase chain reaction (qRT–PCR) was used to amplify lyssavirus RNA extracted from bat brain tissue using TRIzol reagent (Invitrogen), following manufacturer's instructions. The assay uses pan-lyssavirus primers and virus-specific probes that allow rapid differentiation between classical rabies virus (RABV), EBLV-1 and EBLV-2 [23]. PCR products from any sample suspected to contain a lyssavirus species other than the three stated were run on an agarose gel using electrophoresis to determine the presence or absence of a specific band. In addition, positive samples were tested using a pan-lyssavirus hemi-nested RT–PCR. Briefly, cDNA was synthesized by reverse transcription, using 2 µl of total RNA, in a mastermix, including MMLV-RT enzyme and a pan-lyssavirus primer (JW12). Two successive amplifications were undertaken using two rounds of PCR, comprising three primers designed to bind to well-conserved regions within the nucleoprotein gene (N gene) [24].
Sequencing and phylogenetic analysis
Amplicons generated by hemi-nested RT–PCR [partial nucleoprotein gene, 606 base pairs (bp)] were cleaned up using Agencourt® AMPure® (Beckman Coulter, High Wycombe, UK) solid phase reversible immobilization, performed on a Biomek NXP Laboratory Automation Workstation (Beckman Coulter, High Wycombe, UK). Purified PCR products were Sanger sequenced using primers JW12 and JW6UNI (or JW10UNI if 1st round negative) and BigDye Terminator v3.1 Cycle Sequencing Kit chemistry. The sequencing products were purified using Agencourt® CleanSEQ® as above. The products were sequenced on ABI 3130xl Genetic Analyser or AB 3730 DNA Analyser (Applied Biosystems/Life Technologies, Paisley, UK). The DNASTAR Lasergene Core Suite was used to align the contigs and make 405 bp consensus sequences. Sequence alignments and Maximum-likelihood phylogenetic trees were generated in MEGA6 [25] with bootstrap replications of 1000 [26].
Geographical analysis of submissions
Of 10 656 bats submitted between 2005 and 2015, 1037 submissions had missing or incorrect geo-referencing data. These missing data were cleansed using Microsoft Excel and ArcGIS and aggregated to a bespoke county level using a combination of partial postcodes, town names and historic county information on the submission form. Due to missing spatial information, 420 submissions were unable to be geo-referenced at all and are therefore not included in the analysis. In order to maximize the number of submissions to be visualized, a composite shapefile of vice counties, ceremonial counties and unitary authorities was created consisting of 69 spatial units.
RESULTS
Passive surveillance in UK bats between 2005 and 2015
This study describes the passive surveillance of lyssaviruses in bats received and tested between 2005 and 2015, contextualizing the previous report covering the period 1987–2004 [20]. In the 2005–2015 study period, 10 656 bats were received and 6891 were tested. Bats submitted but not tested, were either pipistrelle species that have had no known contact with humans or other animals (2953), or untestable carcasses (812), for example desiccated or decomposed. This is relatively common because of the ad hoc discovery of carcasses, often found outdoors a considerable period of time after death.
All bats tested were reported as negative for lyssavirus infection with the exception of seven Daubenton's bats all of which tested positive for EBLV-2 (Table 2). Two further EBLV-2 positive Daubenton's bats were identified in 2016 [27] (data from 2016 passive surveillance is not formally included in this report as the data were not complete at the time of analysis), totalling 9 EBLV-2 cases diagnosed through passive surveillance in the UK Daubenton's bat population since 2005 and 13 cases since passive surveillance began. A saliva swab collected through active surveillance in Scotland in 2008 was PCR positive and confirmed by sequencing to be EBLV-2, although no virus has been isolated. Additionally, a human case was diagnosed in 2002, totaling 15 EBLV-2 infections across the UK since records began (Table 2). No other lyssavirus species have been identified in bats in the UK, and no bat species other than M. daubentonii has been identified with a lyssavirus infection.
Table 2.
Details of reported EBLV-2 cases within the United Kingdom, including public health risk factors
| Year | Case reference | Location | Type of location | Sample received | Species | Sex | Age | Human contact? | Animal contact? | Reference | Genbank accession number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1996 | RV628 | New Haven, Sussex, England | Private premises | 30/05/1996 | M. Daubentonii | Female | Adult | Bite | nk | Whitby et al. [40] | U89478 |
| 2002 | RV1332 | Carnforth, Lancashire, England | Private premises | 07/07/2002 | M. Daubentonii | Female | Juvenile | Cared for | Cat | Johnson et al. [41] | AY212120 |
| 2002 | RV1333 | Angus, Scotland | na | 11/11/2002 | Human | Male | Adult (55 years) | na | nk | Fooks et al. [9] | EF157977 |
| 2003a | RV1788 | Blackburn, Lancashire, England | Public area | 26/10/2004 | M. Daubentonii | Male | Adult | Cared for | nk | Fooks et al. [17] | JQ796808 |
| 2004 | RV1787 | Staines, Surrey, England | Public area | 28/09/2004 | M. Daubentonii | Female | Juvenile | Cared for | nk | Fooks et al. [17] | JQ796807 |
| 2006 | RV2159 | Abingdon, Oxfordshire, England | Public area | 12/09/2006 | M. Daubentonii | Female | Juvenile | Cared for | nk | Fooks et al. [42] | JQ796809 |
| 2007 | RV2336 | Stokesay Castle, Shropshire, England | Tourist attraction | 12/08/2007 | M. Daubentonii | Female | Adult | Bite | nk | Harris et al. [28] | JQ796810 |
| 2008 | RV2418 | Teddington, Surrey, England | Public area | 07/05/2008 | M. Daubentonii | Female | Adult | Cared for and used as show bat | Cat | Pajamo et al. [43] | JQ796811 |
| 2008 | RV2473 | Stokesay Castle, Shropshire, England | Tourist attraction | 25/09/2008 | M. Daubentonii | Male | Juvenile | nk | nk | Banyard et al. [29] | JQ796812 |
| 2008b | M08/09 | Forteviot, Perthshire | nk | 14/07/2008 | M. Daubentonii | Male | Adult | nk | nk | McElhinney et al. [8] | JQ796804 |
| 2009 | RV2482 | Linlithgow Palace, West Lothian, Scotland | Tourist attraction | 23/08/2009 | M. Daubentonii | Female | Adult | nk | nk | Horton et al. [44] | JQ796806 |
| 2014 | RV2974 | Stokesay Castle, Shropshire, England | Tourist attraction | 22/07/2014 | M. Daubentonii | Undetermined | Undetermined | nk | nk | Unpublished | KY688148 |
| 2015 | RV3158 | Newtown, Powys, Wales | Private premises | 14/07/2015 | M. Daubentonii | Male | Juvenile | nk | nk | Unpublished | KY688153 |
| 2016 | RV3369 | Bolton Priory Church, Yorkshire | Tourist attraction | 10/08/2016 | M. Daubentonii | Female | Juvenile | Cared for | nk | Johnson et al. [27] | KY688156 |
| 2016 | RV3370 | Haydon Bridge, Northumberland | Public area | 02/09/2016 | M. Daubentonii | Male | Adult | Cared for | nk | Johnson et al. [27] | KY688155 |
Carcass frozen and submitted for testing October 2004.
EBLV-2 RNA detected in an oral swab taken as part of surveillance for lyssaviruses in Scotland. na, not applicable; nk, not known.
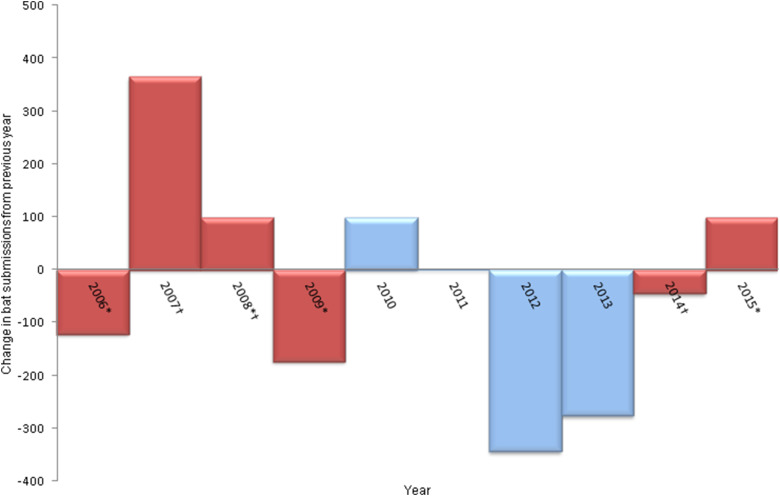
The annual number of submissions ranged from 571 (2014) to 1309 (2008), with a mean of 969 bats (Table 1). The change in annual submissions ranged from −344 (between 2011 and 2012) to +365 (2006 and 2007, Fig. 1), with a mean change of −30 bats (standard deviation of the mean 198·8) across the study period. Every year (except one) following an EBLV-2 case (2007, 2008, 2010 and 2015) an increase in the total number of bat submissions is observed. The exception is between 2008 and 2009, during which there was a reduction of 174 submissions. Between 2010 and 2013, no positive cases were identified, and each subsequent year submissions remained stable or decreased. This trend suggests that the variation in the number of bats submitted is related to the awareness of EBLV-2 in the UK, which is often heightened in the period following the identification of a new positive case.
Table 1.
Number of each bat species submitted 2005–2015
| Year received | Pipistrelle sp. | Plecotus sp. | M.mys/M.bra | M.dau | M.nat | R.hip | E.Ser | N.noc | N.lei | R.fer | B.bar | M.bec | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | 748 | 123 | 42 | 17 | 28 | 12 | 10 | 6 | 2 | 0 | 1 | 0 | 2 | 991 |
| 2006 | 653 | 125 | 34 | 23 | 16 | 6 | 10 | 2 | 8 | 1 | 0 | 1 | 2 | 881 |
| 2007 | 934 | 166 | 39 | 28 | 21 | 4 | 8 | 8 | 4 | 1 | 4 | 0 | 2 | 1219 |
| 2008 | 955 | 166 | 33 | 70 | 36 | 19 | 15 | 8 | 5 | 0 | 0 | 1 | 1 | 1309 |
| 2009 | 853 | 153 | 35 | 22 | 20 | 14 | 8 | 8 | 4 | 1 | 0 | 0 | 6 | 1125 |
| 2010 | 927 | 132 | 49 | 24 | 20 | 18 | 7 | 3 | 5 | 1 | 2 | 0 | 4 | 1192 |
| 2011 | 953 | 139 | 32 | 29 | 25 | 15 | 3 | 10 | 6 | 3 | 1 | 1 | 1 | 1219 |
| 2012 | 583 | 194 | 33 | 31 | 23 | 13 | 6 | 2 | 2 | 1 | 1 | 3 | 7 | 899 |
| 2013 | 370 | 125 | 19 | 19 | 16 | 15 | 8 | 6 | 2 | 3 | 1 | 1 | 6 | 592 |
| 2014 | 370 | 112 | 16 | 26 | 22 | 5 | 8 | 5 | 2 | 3 | 0 | 2 | 0 | 571 |
| 2015 | 419 | 132 | 21 | 35 | 27 | 6 | 6 | 7 | 1 | 0 | 2 | 1 | 1 | 658 |
| Total | 7765 | 1567 | 353 | 324 | 254 | 127 | 89 | 65 | 41 | 14 | 12 | 10 | 32 | 10 656 |
| Mean (per year) | 706 | 142 | 32 | 29 | 23 | 12 | 8 | 6 | 4 | 1 | 1 | 1 | 3 | 969 |
| Estimated UK population | 2 000 000 | 201 000 | 70 000 | 150 000 | 100 000 | 14 000 | 15 000 | 50 000 | 10 000 | >4000 | 5000 | 1500 | NA | 2 666 500 |
Estimated UK populations are taken from Harris et al. [14]. Pipistrellus species (sp.) includes Pipistrellus pipistrellus (P. pip), Pipistrellus pygmaeus (P. pyg), Pipistrellus nathusii (P. nat), Pipistrellus kuhlii (P. kul) and specimens unidentifiable to species level. Plecotus sp. includes Plecotus auritus (Pl. aur), Plecotus austriacus (Pl. aus) and specimens unidentifiable to species level. NA, not applicable. Species nomenclature: B.bar, Barbastella barbastellus; E.ser Eptesicus serotinus; M.bec, Myotis bechsteinii; M.dau, Myotis daubentonii; M.mys, Myotis mystacinus; M.bra, Myotis brandtii; M.nat, Myotis nattererii; N.lei Nyctalus leisleri; N.noc, Nyctalus noctula; R.fer, Rhinolophus ferrumequinum; R.hip, Rhinolophus hipposideros.
Fig. 1.
Annual change in the number of bat submissions. A red bar denotes that at least one EBLV-2 positive bat was identified during that year. †Positive case from Stokesay Castle, *positive case from elsewhere in the UK.
Bat submissions by species and location
Eighteen species of bats were submitted between 2005 and 2015, 16 resident to the UK, and two considered non-native vagrants [five Kuhl's pipistrelle (Pipistrellus kuhlii) and one parti-coloured bat (Vespertilio murinus)]. Of the five Kuhl's pipistrelles, four were submitted from Jersey between 2007 and 2010. The remaining Kuhl's pipistrelle was found in St. Pancreas London underground station in 2006. All five were identified by a bat specialist (A.M. Hutson, personal communication). The Parti-coloured bat was found on the Isle of Arran (Scotland) in 2011.
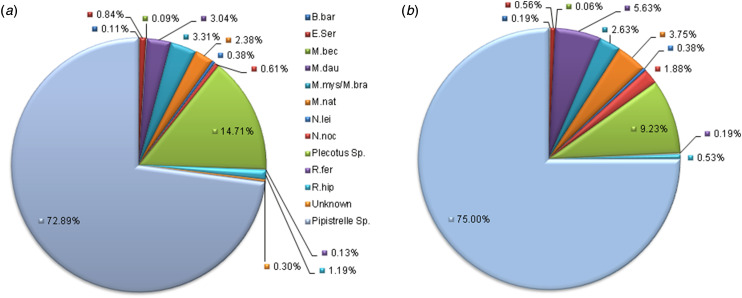
The only resident bat species not received was Alcathoe's bat (M. alcathoe), which is difficult to distinguish morphologically from the whiskered bat (M. mystacinus) and Brandt's bat (M. brandtii) and therefore it is possible that some submissions identified as the more common myotis species, might be Alcathoe's bat. Speciation using mitochondrial cytochrome b gene specific PCR is required to confidently distinguish these three species but this is not part of the current surveillance scheme. The majority of submissions were of native pipistrelles (Pipistrellus sp.) and long-eared bats (Plecotus sp.) which account for 72·9% (7765 specimens) and 14·7% (1567 specimens), respectively. A total of 324 Daubenton's bats were submitted, comprising 3% of total submissions (Fig. 2a), of which 277 were tested and seven (2·5% Daubenton's bats tested) were positive for EBLV-2 during the period 2005–2015. If surveillance was uniform across species, we would expect to see rates of submission similar to their relative population sizes [14,20] (Fig. 2b). In general, submission rates are relative to the population size, with Pipistrellus sp. representing 73% of actual submissions compared with 75% of expected submissions. However, the lesser horseshoe bat (Rhinolophus hipposideros) is over represented, with approximately twice the expected percentage submitted, conversely, the noctule (Nyctalus noctula) is under represented (Fig. 2).
Fig. 2.
(a) Percentage of total bat submissions per species 2005–2015. M.mys and M.bra are grouped as it is not possible to confidently separate them morphologically. (b) Expected percentage of total bat submissions per species, based on UK population estimates [14].
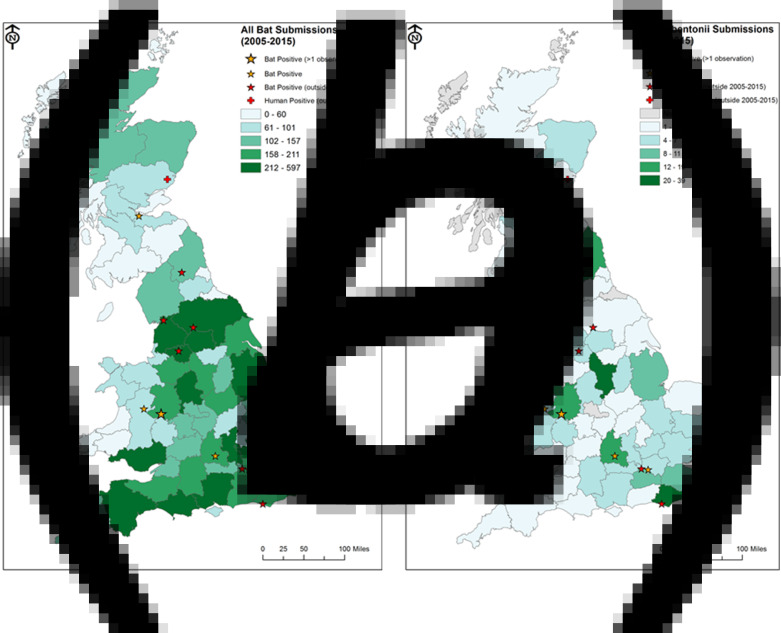
Bats were received from all but one of the counties within the UK. The highest numbers of submissions were received from Kent, Lancashire and Buckinghamshire (5·6%, 4% and 3·8% of submissions, respectively, Fig. 3a). It was not possible to attribute 3·9% of submissions to any county therefore these were removed from the geographic analysis. The number of submissions received from individual counties was highly variable and ranged from 1 to 597. The most common range of submissions from a county was 121–140 bats (eight counties). One county (Orkney) did not submit any bats during the 11 years; however, only one species of bat (P. pipistrellus) breeds on the Orkney Islands and only at a very small number of sites. Three counties submitted more than 400 bats. This indicates that there is considerable spatial variation in submission rate.
Fig. 3.
Submissions of (a) total bats, and (b) Daubenton's bats, to the passive surveillance scheme, 2005–2015. Stokesay Castle is indicated as the only site with more than 1 positive case reported (larger yellow star).
Submissions of Daubenton's bats
Daubenton's bats were received from 60 of the 69 counties within the UK. Where bats were submitted the number ranged from 1 to 39, with the majority of counties (n = 46) submitting between 1 and 5 Daubenton's bats during the 11-year period. Nine counties did not submit any Daubenton's bats, highlighting gaps in surveillance across the UK; particularly in Scotland, which includes five of the nine counties (Fig. 3b). However, the distribution of Daubenton's is not uniform across the country; therefore the lack of submissions from these counties could be a reflection of this diversity. Only two counties submitted over 30 Daubenton's bats. In contrast with overall submissions, the highest numbers of Daubenton's bat submissions were from Derbyshire (12% of the total), East Sussex (10·2%) and Shropshire (5·9%). Thirty-nine Daubenton's bats were received from Derbyshire between 2005 and 2015; however, 25 of these were made in a single submission. As seen for bat submissions as a whole, there is substantial spatial variation in submissions of Daubenton's bats across the UK despite their broad geographical range.
Submissions of bats from Stokesay Castle
During the study period, the annual number of submissions of bat carcasses of any species from Shropshire have progressively increased over time since the first submission in 1998; with 14 submitted up to 2004 then between 10 and 24 submissions annually (excepting 2012 when seven were received). The annual submissions from Shropshire range between 1% (n = 13/1309 in 2008) and 3% (n = 20/658 in 2015) of the national total submitted each year, with a total of 1·6% of all bats submitted between 2005 and 2015 coming from this county (Fig. 3b).
Stokesay Castle, in Shropshire (latitude 52·4281°N, longitude 2·8298°W), is a medieval fortified manor house open to the public as a visitor attraction and is known to host a maternity roost for Daubenton's bats, thought to exceed a count of 100 bats at peak periods. Staff at the site undertake a daily inspection for grounded bats in the summer, and those that die are identified, and submitted to APHA for testing. Since 2007, a total of 21 bats have been submitted from Stokesay Castle, with annual submissions ranging between 1 and 3 bats per year, comprising 15 Daubenton's bats, five Pipistrellus sp. and one Natterer's bat. The first recorded submission in 2007 tested positive for EBLV-2 [28]. Additional cases of EBLV-2 in Daubenton's bats from the site were confirmed in September 2008 [29] and then in July 2014. The 2014 case was the 10th confirmed case of EBLV-2 in a British Daubenton's bat and identified Stokesay Castle as the only site in Europe to have recorded more than one case.
Location and exposure in EBLV-2-positive cases
All 13 EBLV-2 passive surveillance cases have been found at locations that can be classified into ‘private premises,’ ‘public areas’ or ‘tourist attractions’ (Table 2). Private premises include houses, gardens and places of work; public areas include parks, footpaths and roads; and tourist attractions include buildings and areas that are open to the public and attract visitors. Almost 40% of UK EBLV-2 positive bats (n = 5/13) were found at tourist attractions (three at Stokesay Castle, Shropshire; one at Linlithgow Palace, West Lothian; one at Bolton Priory, Yorkshire). Of the remaining eight cases, five were found in public areas. Frequency of contact between bats and humans, and bats and other animals was assessed for the EBLV-2-positive bat cases, using comments and notes from the paperwork received with the specimens. Contact with humans was described in 69% of cases (n = 9/13), either through caring for the bat or through a bite received during handling. This figure is substantially higher than the 4% of total submissions that were recorded as having human contact (data not shown). Contact with domestic cats was recorded in 15% (n = 2/13) of positive cases, compared with 20% of total submissions (which included contact with any other animal).
Phylogenetic analysis of UK EBLV-2 isolates
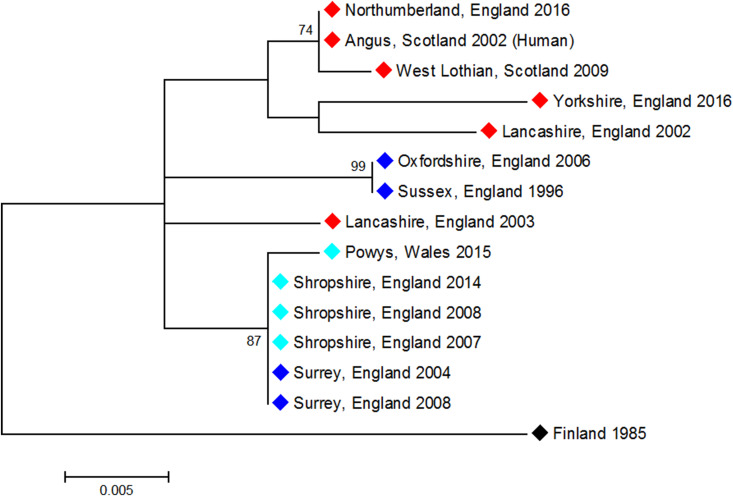
Partial N gene sequences (405 bp) for all known UK EBLV-2 cases were generated, aligned and analysed to determine their phylogeny. Partial N sequence from an EBLV-2 isolate originating from Finland in 1985, is included in the analysis as an outgroup. The Maximum-likelihood method was used to infer phylogenetic relationships in MEGA software [25].
The UK EBLV-2 sequences cluster geographically rather than temporally, albeit that bootstrap values are not always high. However, the viruses isolated from cases found across a central area of Great Britain (including the cases from Shropshire and Wales) form a monophyletic clade along with southern isolates RV1787Surrey2004 and RV2418Surrey2008 with high bootstrap support (Fig. 4, Table 2). This analysis supports the general correlation between geographical location and genetic relatedness shown previously [8], however, in central UK, there are two clades (Shropshire/Surrey and Oxford/Sussex) in overlapping areas. The three cases from Stokesay Castle are identical across the 405 bp region sequenced, despite being isolated 7 years apart.
Fig. 4.
Molecular phylogenetic analysis of partial N gene (405 bp) sequences from UK EBLV-2 cases using the Maximum-likelihood method (MEGA6). Analysis was performed with bootstrap of 1000, values >60 indicated. Colour key is geographic; red, northern UK (including Scotland and North England); light blue, Central UK (including Wales); dark blue, southern UK.
DISCUSSION
Passive surveillance for lyssavirus infection in British bats provides valuable insight into the epidemiology of EBLV-2 in the UK as well as providing decision-makers with information useful for protecting public health. These analyses provide strong support for the presence of endemic circulation of EBLV-2 in Daubenton's bats at the national scale, regional geographic structures within which distinct viruses can be maintained, and the first evidence that individual roosts may maintain infection for prolonged periods. Although the scheme has tested a total of 15 539 bats between 1987 and 2015 its limitations reduce the power of inference that it supports, primarily the reliance on ad hoc voluntary submissions, which inevitably lead to inconsistent temporal and geographical representation. In addition, there has been a decline in overall submissions in the UK since 2011, reducing the breadth and depth of surveillance further.
The number of bats received per species generally reflects the differences in estimated population numbers, with more specimens received for those species considered more common. Representation per species varies with Daubenton's bats being relatively under represented. It must be taken into consideration that population estimates for UK bat species have inherent uncertainty, as they are based either on limited data, or opinion of experts where no data is available [14,30]. Therefore, it is impossible to accurately assess how well represented each species is within the passive surveillance scheme. Furthermore, the low numbers of Daubenton's received are likely to be related to their behaviour and choice of roost sites as they are less synanthropic than other more commonly submitted species (e.g., Pipistrellus pipistrellus). The over representation of synanthropic species has previously been highlighted as a potential cause of bias in passive surveillance schemes [31], because of their increased likelihood of contact with people. Despite these potential biases, the relatively small number of Daubenton's bats submitted compared with the overall population is not necessarily problematic; rather the representativeness of the sample obtained is more relevant. The consistent but sporadic detection of cases over the time period of the study, that are genetically identical over the genomic region analysed, suggests that in the absence of another wildlife reservoir, EBLV-2 infection has persisted in the local Daubenton's bat population for over 7 years. Within the context of the current surveillance programme and its opportunistic sampling, it is only possible to estimate disease incidence at local/roost level where sampling may be considered representative of that roost or local population, as opposed to a national level where sampling has more biases and much less coverage [32]. Stokesay Castle is unique as the only site from which multiple EBLV-2 infected bats have been identified, in spite of sampling at several other roosts of Daubenton's bats over a similar time period. In addition, staff check the site daily for grounded bats, moribund and dead bats, whereas the majority of submissions from other sites are opportunistic findings. Staff follow a set protocol for bat observation and detection and details of bat findings are well documented. Hence, this site provides an opportunity to understand the true site specific infection pattern of EBLV-2 and suggests that the virus can be maintained at a relatively small scale. However, confirming this hypothesis would require greater understanding of movement and mixing among roosts, systematic longitudinal sampling, and detailed information regarding the effective population size both during and between the birthing seasons. If the detections of EBLV-2 infected bats at Stokesay Castle are due to substantial and structured local surveillance, this implies that other sites could have similar persistence of infection that has so far gone undetected, and such sites (such as those with a single previous case of EBLV-2) warrant further investigation. To better understand EBLV-2 infection dynamics in Daubenton's bats at the Stokesay Castle site, implementing a targeted research project here and at other sites of interest (such as Newtown, Powys) to determine prevalence of lyssavirus specific antibodies and excretion of virus in saliva in bat populations would give a better understanding of EBLV-2 at these locations. In addition, it would allow comparison of seroprevalence in Daubenton's bats at Stokesay Castle with that of the UK as a whole, and provide information useful for informing the public health risk. The current study shows that 2·5% of all Daubenton's bats tested between 2005 and 2015 were infected with EBLV-2. When analysing Stokesay castle specifically, 33% of all submitted Daubenton's bats tested positive between 2007 and 2015. A comparison of seroprevalence at EBLV-2 positive sites across the UK would improve the picture of seroprevalence at Stokesay Castle, at other sites of interest and across the UK as a whole.
Lyssaviruses are mostly associated with specific reservoir host species (arguably with the exception of RABV [33]) and only rarely transmit to non-reservoir hosts [34], with ongoing transmission from a spill-over event considered rarer still. This observation likely explains why there are very few reports of EBLV infections in insectivorous bat species other than serotines (EBLV-1), or Daubenton's bats and Pond bats (EBLV-2), despite documented co-roosting with other bat species. It is interesting to note that EBLV-1 was detected in three species (brown long eared bats and two pipistrelle species) in a retrospective study of bats in Germany [35]. Whether infection in these species resulted from spill-over events related to co-roosting with serotine bats is not clear. Undertaking surveillance of other bat species at the Stokesay Castle site is important to gain a clearer picture of infection for those species at close quarters with an infected population of Daubenton's bats.
Our results suggest that the epidemiological unit capable of independently maintaining circulation of the virus may be smaller than previously thought, whether measured by host population size or geographical area. The molecular epidemiology of EBLVs was reviewed by McElhinney et al. in 2013 [8]. Genetic relationships between EBLV-2 isolates were determined using a 400 bp region of the N gene, which is equivalent with the 405 bp region used in this study. EBLV-2 sequences grouped geographically, which is also supported by this study. Previously, only the distinction between isolates from the Scotland and northern England cluster and those from the south of England were possible [8]. Here, the presence of isolates characteristic of a ‘central UK’ clade is added, suggesting a smaller sub-national spatial epidemiological scale within which a distinct isolate can be maintained. The region of the N gene analysed in this study is sufficient to confirm the geographical clustering of UK cases, however, using a more variable region of the genome or full genomic sequence is likely to provide greater resolution and insight into the genetic variation between isolates. It would be interesting to see whether this distinguishes the Shropshire and Powys isolates from the more geographically distant Surrey isolates, and would allow further investigation of the Stokesay castle viruses (which have been isolated over a 7 year period and appear highly related), as well as investigation of the transmission and evolution of a virus within a population. Further investigation of the apparent geographic overlap of genetically distinct strains, combined with information on population dynamics would not only inform the understanding of viral evolution and dynamics, but also any variation in regional public health risk.
The 13 cases of EBLV-2 identified by passive surveillance in UK Daubenton's bats, combined with population data suggesting that Daubenton's bats do not migrate long distances, confirm previous early speculation that EBLV-2 is circulating endemically within the UK [20,29,36], however, the mechanism for how the virus is transmitted and maintained within wild bat communities and populations is still poorly understood [29]. An examination of the genetics of the host [37] indicates that Daubenton's bats have spatial clustering that could help isolate the short-term evolution of the virus, but that some long-distance movements also occur, which would help maintain disease persistence at the continental scale [38].
In general, bat submissions rise following a positive report and decrease following sequential years without a positive report. This observation was previously made for UK bat submissions between 1987 and 2004, during which the annual total submissions were substantially higher following a positive case [31]. Identification of an EBLV-2 case often results in media interest either locally or nationally. These trends in variation of number of bats submitted, do not correlate with the relatively stable trends in population size suggested for many bat species nationally [39]. This may demonstrate a variation in the awareness of EBLV-2 across the general public in the UK, which becomes heightened in the period of time following the identification of a new EBLV-2 case and its local or national reporting. This observation underlines the importance of continued communication and responsible promotion of the scheme to the general public, in addition to maintaining good working relationships with all NGOs, veterinarians and other professional groups which either submit bats or promote the surveillance scheme. In addition, it highlights the indirect but significant effect that the media can have on passive surveillance involving public reporting; continuing to report and publicize individual cases therefore will be important to the on-going success of the scheme. This is exemplified by the recent publication that described the submission and testing of two EBLV-2-positive Daubenton's bats from two separate locations in the UK in 2016 [27]. Importantly, detection of lyssaviruses in bats serves to educate on the risks associated with bat contact. Human or animal infection and death following a spillover event has a damaging effect on the public perception of bats and as such, increasing our understanding of these viruses and their distribution in UK bat populations is of great importance. Efforts to raise awareness of the scheme with the general public and veterinary practitioners, and maintaining ongoing positive interactions and collaborations with bat groups has allowed this uniquely comprehensive surveillance to function thus far, and further engagement is likely to improve the number of submissions and increase the quality of surveillance across the UK. It is important to note that much of the success of the scheme, especially the submission of eventual confirmed EBLV-2 cases has relied on the engagement and interest of informed bat enthusiasts and bat-carers encountering moribund Daubenton's bats or carcasses. We would like to thank them collectively here for continuing to recognize the importance of submitting dead bats to the scheme and supporting the slow but necessary accretion of knowledge that will eventually lead to the better understanding of this disease of public concern. The continuing, sporadic detection of EBLV-2 infected bats in the UK highlights the importance of the continuing passive surveillance, and the Stokesay Castle site has the potential to provide further information on the nature of viral maintenance within a natural population.
ACKNOWLEDGEMENTS
The authors thank Miss Daisy Jennings, Miss Isobel Jarvis and all staff at the APHA Central Sequencing Unit for technical support. Thanks are given to the Bat Conservation Trust, local bat groups, Stokesay castle staff, veterinary staff at the APHA Field Services Offices and the general public for taking the time to submit bats for testing.
This work was supported by the Department for Environment Food and Rural Affairs (Defra); Scottish Government and Welsh Government (grant number SV3500).
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001455.
click here to view supplementary material
DECLARATION OF INTEREST
Dr Andrew Breed acts as an editor for Epidemiology and Infection. Dr Anthony R. Fooks was a previous editor for Epidemiology and Infection (2006–2014) and is now a member of the editorial board for the journal. The remaining authors declare no conflicts of interest.
REFERENCES
- 1.ICTV virus taxonomy: 2015 release (http://www.ictvonline.org/virustaxonomy.asp). Accessed 15 November 2016.
- 2.Ceballos NA, et al. Novel lyssavirus in bat, Spain. Emerging Infectious Diseases 2013; 19: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunawardena PS, et al. Lyssavirus in Indian flying foxes, Sri Lanka. Emerging Infectious Diseases 2016; 22: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans JS, et al. Rabies virus vaccines: is there a need for a pan-lyssavirus vaccine? Vaccine 2012; 30: 7447–7454. [DOI] [PubMed] [Google Scholar]
- 5.Banyard AC, et al. Lyssaviruses and bats: emergence and zoonotic threat. Viruses 2014; 6: 2974–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Racey PA, Hutson AM, Lina PH. Bat rabies, public health and European bat conservation. Zoonoses Public Health 2013; 60: 58–68. [DOI] [PubMed] [Google Scholar]
- 7.Hammarin AL, et al. Lyssavirus-reactive antibodies in Swedish bats. Infection Ecology & Epidemiology 2016; 6: 31262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McElhinney L, et al. Molecular epidemiology of bat lyssaviruses in Europe. Zoonoses Public Health 2013; 60: 35–45. [DOI] [PubMed] [Google Scholar]
- 9.Fooks AR, et al. Case report: isolation of a European bat lyssavirus type 2a from a fatal human case of rabies encephalitis. Journal of Medical Virology 2003; 71: 281–289. [DOI] [PubMed] [Google Scholar]
- 10.Lumio J, et al. Human rabies of bat origin in Europe. Lancet 1986; 1: 378. [DOI] [PubMed] [Google Scholar]
- 11.Daoust PY, Wandeler AI, Casey GA. Cluster of rabies cases of probable bat origin among red foxes in Prince Edward Island, Canada. Journal of Wildlife Diseases 1996; 32: 403–406. [DOI] [PubMed] [Google Scholar]
- 12.Leslie M, et al. Bat-associated rabies virus in skunks. Emerging Infectious Diseases 2006; 12: 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos A, et al. Susceptibility of ferrets (Mustela putorius furo) to experimentally induced rabies with European bat lyssaviruses (EBLV). Journal of Veterinary Medicine B, Infectious Diseases and Veterinary Public Health 2004; 51: 55–60. [DOI] [PubMed] [Google Scholar]
- 14.Harris S, et al. A Review of British Mammals: Population Estimates and Conservation Status of British Mammals Other than Cetaceans. Peterborough: The Joint Nature Conservation Committee, 1995. [Google Scholar]
- 15. Daubenton's bat trends for Great Britain (http://www.bats.org.uk/pages/-daubentons_bat-815.html). Accessed 16 November 2016.
- 16.Croft S, Chauvenet ALM, Smith GC. A systematic approach to estimate the distribution and total abundance of British mammals. PLoS ONE 2017; 12: e0176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fooks AR, et al. Detection of antibodies to EBLV-2 in Daubenton's bats in the UK. The Veterinary Record 2004; 154: 245–246. [PubMed] [Google Scholar]
- 18.Brookes SM, et al. European bat lyssavirus in Scottish bats. Emerging Infectious Diseases 2005; 11: 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris SL, et al. Targeted surveillance for European bat lyssaviruses in English bats (2003–06). Journal of Wildlife Diseases 2009; 45: 1030–1041. [DOI] [PubMed] [Google Scholar]
- 20.Harris SL, et al. Passive surveillance (1987 to 2004) of United Kingdom bats for European bat lyssaviruses. The Veterinary Record 2006; 159: 439–446. [DOI] [PubMed] [Google Scholar]
- 21.Dean DJ, Abelseth MK. Laboratory techniques in rabies: the fluorescent antibody test. Monograph Series World Health Organization 1973; 23: 73–84. [PubMed] [Google Scholar]
- 22.Webster WA. A tissue culture infection test in routine rabies diagnosis. Canadian Journal of Veterinary Research 1987; 51: 367–369. [PMC free article] [PubMed] [Google Scholar]
- 23.Wakeley PR, et al. Development of a real-time, TaqMan reverse transcription-PCR assay for detection and differentiation of lyssavirus genotypes 1, 5, and 6. Journal of Clinical Microbiology 2005; 43: 2786–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heaton PR, et al. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. Journal of Clinical Microbiology 1997; 35: 2762–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenstein J. Confidence limits on phylogenies: an Approach Using the Bootstrap. Evolution 1985; 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 27.Johnson N, et al. Two EBLV-2 infected Daubenton's bats detected in the north of England. Veterinary Record 2016; 179: 311–312. [DOI] [PubMed] [Google Scholar]
- 28.Harris SL, et al. Isolation of European bat lyssavirus type 2 from a Daubenton's bat (Myotis daubentonii) in Shropshire. The Veterinary Record 2007; 161: 384–386. [DOI] [PubMed] [Google Scholar]
- 29.Banyard AC, et al. Repeated detection of European bat lyssavirus type 2 in dead bats found at a single roost site in the UK. Archives of Virology 2009; 154: 1847–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russ JM. The Microchiroptera of Northern Ireland: community composition, habitat associations and ultrasound, Thesis. Belfast, Ireland: Queen's University, 1999. [Google Scholar]
- 31.Harris SL, et al. European bat lyssaviruses: distribution, prevalence and implications for conservation. Biological Conservation 2006; 131: 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith GC, et al. EBLV-2 prevalence in the United Kingdom as determined by surveillance testing. Developments in Biologicals 2006; 125: 265–271. [PubMed] [Google Scholar]
- 33.Streicker DG, et al. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 2010; 329: 676–679. [DOI] [PubMed] [Google Scholar]
- 34.Mollentze N, Biek R, Streicker DG. The role of viral evolution in rabies host shifts and emergence. Current Opinion in Virology 2014; 8: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatz J, et al. Enhanced passive bat rabies surveillance in indigenous bat species from Germany – a retrospective study. PLoS Neglected Tropical Diseases 2014; 8: e2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson N, et al. European bat lyssavirus type 2 RNA in Myotis daubentonii. Emerging Infectious Diseases 2006; 12: 1142–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atterby H, et al. Population genetic structure of the Daubenton's bat (Myotis daubentonii) in western Europe and the associated occurrence of rabies. European Journal of Wildlife Research 2010; 56: 67–81. [Google Scholar]
- 38.Hagenaars TJ, Donnelly CA, Ferguson NM. Spatial heterogeneity and the persistence of infectious diseases. Journal of Theoretical Biology 2004; 229: 349–359. [DOI] [PubMed] [Google Scholar]
- 39.National bat monitoring programme annual report 2015 (http://www.bats.org.uk/pages/nbmp_annual_report.html). Accessed 10 February 2017.
- 40.Whitby JE, et al. First isolation of a rabies-related virus from a Daubenton's bat in the United Kingdom. Veterinary Record 2000; 147: 385–388. [DOI] [PubMed] [Google Scholar]
- 41.Johnson N, et al. Isolation of a European bat lyssavirus type 2 from a Daubenton's bat in the United Kingdom. Veterinary Record 2003; 152: 383–387. [DOI] [PubMed] [Google Scholar]
- 42.Fooks AR et al. Isolation of EBLV-2 in a Daubenton's bat (Myotis daubentonii) found in Oxfordshire. Veterinary Record 2006; 159: 534–535. [DOI] [PubMed] [Google Scholar]
- 43.Pajamo K, et al. Isolation of European bat lyssavirus type 2 (EBLV-2) in a Daubenton's bat in the UK with a minimum incubation period of 9 months. Rabies Bulletin Europe 2008; 32: 6–7. [Google Scholar]
- 44.Horton DL, et al. European bat lyssavirus type 2 in a Daubenton's bat in Scotland. Veterinary Record 2009; 165: 383–384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001455.
click here to view supplementary material