Figure 3.
Interaction of SARS-CoV, SARS-CoV-2, MERS-CoV, EBOV, and NiV with their receptors
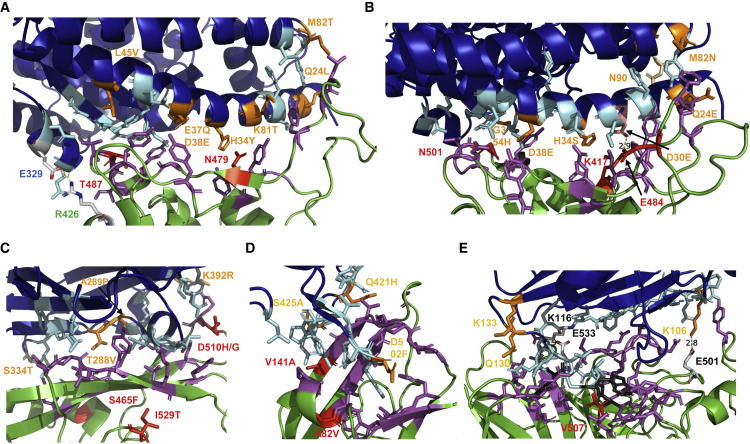
(A) Interaction surface between ACE2 and S of SARS-CoV: ACE2 is colored blue, S in green. The contacting amino acids are shown as cyan (ACE2) or magenta (S) sticks. The seven amino acids substituted in civet ACE2 are labeled in orange. In ACE2 of mice, M82 is substituted to N, thereby creating an N-glycosylation site. The two amino acid substitutions during adaptation of SARS-CoV from civets to humans (N479) and from humans to humans (T487) are shown in red. Glu329 in S and Arg426 in ACE2 form a salt bridge at the periphery of the interaction surface. This figure was generated with PyMol 2.1.1. using the pdb file 2AJF. The contacting amino acids shown here are from Lan et al. (2020), which differ in four peripheral residues from the first publication Li et al. (2005a).
(B) Interaction surface between ACE2 and S of SARS-CoV-2: ACE2 is colored blue, S in green. The contacting amino acids are shown as cyan (ACE2) or magenta (S) sticks. The seven amino acids substituted in pangolin ACE2 are labeled in orange. Lys417 in S and Glu30 in ACE2 form a salt bridge in the middle of the interaction surface. Lys417 (labeled red) is substituted by a Thr in variant P1 and by an Asn in variant B 1.351. Highlighted as a red stick is also N501, which is substituted by Tyr in variants B1.1.1, B1.351. and P1. The figure was generated with PyMol 2.1.1. using the pdb file 6M0J.
(C) Interaction surface between human DPP4 and S of MERS-CoV: DPP4 is colored blue, S in green. The contacting amino acids are shown as cyan (DPP4) or magenta (S) sticks. The four amino acids substituted in camel DPP4 are labeled in orange. Residue 334 is not directly contacting the spike but is an N-glycosylation site in mice DPP4, which needs to be removed to make mice susceptible to MERS-CoV infection. The two amino acid substitutions (S465F, D510H) during adaptation of S to a suboptimal bat-associated virus receptor and during MERS outbreak in South Korea (I529T, D510G) are shown in red. The figure was generated with PyMol 2.1.1. using the pdb file 4KRO.
(D) Interaction surface between human NPC1 and GP of EBOV. NPC1 is colored blue, GP in green. The contacting amino acids are shown as cyan (NPC1) or magenta (GP) sticks. The three amino acids substituted in pig NPC1 are labeled in orange. Amino acid substitutions restoring EBOV binding to refractory receptors (V141A) and during the large Ebola epidemic (A82V) are labeled red. The figure was generated with PyMol 2.1.1. using the pdb file 5F1B.
(E) Interaction surface between human ephrin-b2 and G of Nipah virus. Ephrin-B2 is colored blue, G in green. Most of the contacting amino acids are shown as sticks. Three hydrophobic amino acids (F120, L124, W125) in the loop of ephrin-B2 essential for binding are colored in gray. The two salt bridges (K106 with E501, K116 with E533) are indicated. The only amino acid substitution between G of Nipah and Hendra virus is shown in red. Receptor amino acids near the binding site variable between species (K106, Q130, K133) are labeled in orange. The figure was generated with PyMol 2.1.1. using the pdb file 2VSM.