Abstract
Normal cells explore multiple states to survive stresses encountered during development and self-renewal as well as environmental stresses such as starvation, DNA damage, toxins or infection. Tumor cells co-opt normal stress mitigation pathways to survive stresses that accompany tumor initiation, progression, metastasis and immune evasion. Cancer therapies accentuate tumor cell stresses and invoke rapid non-genomic stress mitigation processes that maintain cell viability and thus represent key targetable resistance mechanisms. In this review, we describe mechanisms by which tumor ecosystems, including cancer cells, immune cells, and stroma, adapt to therapeutic stresses and describe three different approaches to exploit stress mitigation processes: a) interdict stress mitigation to induce cell death, b) increase stress to induce cellular catastrophe and c) exploit emergent vulnerabilities in tumor and microenvironment cells. We review challenges associated with tumor heterogeneity, prioritizing actionable adaptive responses for optimal therapeutic outcomes, and development of an integrative framework to identify and target vulnerabilities that arise from adaptive responses and engagement of stress mitigation pathways. Finally, we discuss the need to monitor adaptive responses across multiple scales and translation of combination therapies designed to take advantage of adaptive responses and stress mitigation pathways to the clinic.
General introduction.
Tumor cells as well as cells in the tumor microenvironment are subjected to stresses inflicted by altered cell states associated with tumor initiation, progression, metastases and immune evasion13. To survive and adapt to these stressors, tumors co-opt endogenous stress mitigation pathways that have evolved to allow normal cells to deal with stresses that occur during development and renewal or as a consequence of environmental perturbations such as those induced by starvation, DNA damage, dietary toxins, infection or cancer therapy4,5. Examples of stresses that tumor cells are subjected to during tumor initiation, progression and metastasis are shown in Figure 1A and discussed in more detail by Luo et al6. Tumor cells, as well as cells in the tumor microenvironment, must also rapidly implement additional mitigation strategies (Figure 1A) to survive stress created by cancer therapies. Radiation, chemo- and targeted therapies, as well as immunotherapies, augment ongoing oncogenic stress, resulting in further dependence on stress mitigation pathways. The ability of tumor cells and tumor ecosystems to co-opt endogenous stress mitigation pathways and heterotypic cell-cell signaling that maintain tissue homeostasis, constitutes key targetable mechanisms of therapeutic resistance (three different therapeutic approaches to exploit these mechanisms are shown in Figure 1B)6,7. Table 1 lists examples of stresses induced by cancer therapies as well as the processes that cellular stress sensors engage to mitigate these stresses.
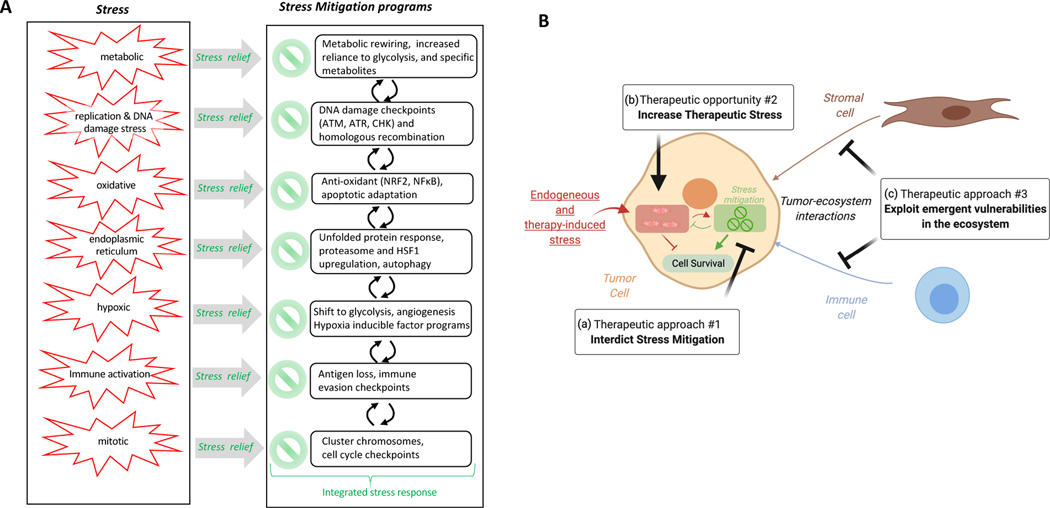
Figure 1: Therapeutic opportunities created by adaptive responses due to tumor cell endogenous stress, engagement of stress mitigation pathways and interactions in the tumor ecosystem.
(A) Stresses associated with tumor initiation and stress mitigation programs that are engaged to allow cancer cells to survive and proliferate. (B) Adaptation to therapeutic stress and engagement of stress mitigation pathways in the tumor ecosystem (tumor, stromal and immune cells) provide three therapeutic options: a) induce tumor cell death by interdicting stress mitigation pathways that allow tumor cell survival, b) overwhelm the mitigation pathways by increasing existing stress that results in stress-induced catastrophe, c) exploit emergent vulnerabilities in the tumor ecosystem by targeting tumor-host interactions. The difference in stress levels and stress mitigation programs between normal and tumor cells provides an opportunity for increased therapeutic index.
Table 1.
Stresses and stress mitigation pathways that can limit therapeutic activity.
| Endogenous and therapeutic stresses | Master regulators of stress mitigation pathways | Stress mitigation pathways | Examples of therapeutic response affected by stress mitigation pathways | Ref |
|---|---|---|---|---|
| Metabolic | AMPK, SIRT1, mTOR, PHD/HIF system | Oxphos/Glycolysis balance, autophagy, mitophagy, mitochondial metabolism, lipid metabolism | Metformin, BRAFi, phenformin | 73,201,202 |
| Oxidative | NRF2, NF-kB, HIF1 | Antioxidant programs, metabolic reprograming | Radiation therapy, temozolomide, gemcitabine | 77,203–205 |
| Apoptosis | p53, TRAIL | BCL2 family balance, epithelial to mesenchymal transition, senescence phenotype | Doxorubicin, paclitaxel, BCL2i, BCL2/XLi, radiation therapy | 206–209 |
| Endoplasmic reticulum/proteostatic | GRP78, PERK, IRE1a, ATF6 | Unfolded protein response, E-Rassociated degradation, heat shock response, proteasome upregulation, autophagy | Chemotherapy, tamoxifen, multi-RTKi | 210 |
| Hypoxia | HIFs | Hypoxic response, angiogenesis, metabolic reprograming, autophagy, extracellular matrix remodelling, immune suppression | anti-VEGFR, multi-RTKi | 211–213 |
| DNA damage/replication stress | ATM, ATR, CHK1, CHK2, RPA, H2AX | Cell cycle checkpoints, DNA repair, anti-apoptotic response, replication stress response | Gemcitabine, PARPi, doxorubicin, cisplatin, radiation therapy, | 214–216 |
| Immune activation | Sting, STAT3, TGFb | Reduced antigen presentation, immune checkpoint activation | Radiation therapy, PARPi, anti-PD1, anti-PD-L1 | 68,217,218 |
Several reviews have covered intrinsic drug resistance mechanisms, including pre-existing tumor clones8,9, as well as adaptive resistance mechanisms, including non-genetic tumor cell state plasticity, microenvironment mechanisms10–16 and drug target-focused mechanisms (drug efflux, drug inactivation, or alterations in drug targets17–22). In this review, we focus on adaptive responses to cancer therapy. These responses represent a unique form of non-genomic resistance that occurs rapidly and is highly reversible and targetable. Importantly, the ability of cells to adapt to the immediate effects of therapeutic stress can facilitate development of irreversible acquired genetic resistance including acquisition of new mutations23–29 or allow selection of pre-existing drug resistant clones30,31. Thus, therapeutic targeting of adaptive resistance provides an opportunity to improve patient outcomes by both immediate inhibition of tumor cell survival, and also by delaying or preventing emergence of genetically resistant clones. However, in order for patients to realize benefit from targeting stress mitigation responses, it is necessary to develop facile approaches to identify the suite of mitigation strategies utilized by individual cells as well as to quantify stress phenotypes and mitigation strategies as they change longitudinally under therapeutic stress. This review highlights examples of stress responses associated with cancer therapies to illustrate concepts that can be generalized across the spectrum of adaptive response and stress mitigation pathways. In addition, we describe three different approaches (Figure 1B) to exploit stress mitigation processes to improve the effectiveness of cancer therapy. Lastly, we discuss opportunities and challenges in implementing these approaches in the clinic. Due to space limitations, the review is designed to provide a framework for broad implementation of therapeutic approaches to capitalize on adaptive responses and engagement of stress mitigation pathways rather than a comprehensive coverage of research and clinical activities.
Section 1: Tumor intrinsic adaptive response programs
Understanding the nature of stresses and adaptive mechanisms (Table 1) that maintain survival of drug-treated tumor cells promises to uncover therapy-induced vulnerabilities and provide rational approaches to target adaptive responses to therapeutic stress to enhance cancer cell death.
Section 1.1: Adaptive responses involving rewiring of signaling pathways
Adaptive responses to signal transduction inhibitors as a special case of stress mitigation:
Given the key role of signaling pathways in the development and progression of cancer, major efforts have been deployed to develop targeted therapies that disrupt pro-oncogenic signaling. Unfortunately, the benefit from such drugs is often temporary due to the ability of cells to rapidly adapt through reactivation or bypass of the targeted molecule, allowing cell survival. In normal cells, signaling pathways have a high level of plasticity leading to stable regulatory networks that result in bistable states with cell populations maintaining distinct phenotypes independently of genotype. This plasticity is built on the balance of negative and positive regulators, availability of alternative isoforms, feedback and feedforward loops and crosstalk with other signaling pathways that are required to maintain normal cellular homeostasis. This plasticity, pathway rewiring, and rapid acquisition of adaptive responses allow tumor cells to mitigate stresses caused by inhibition of signaling mediators by targeted therapies. Pathway bypass through signaling rewiring does not engage a “professional” stress mitigation pathway but rather co-opts the normal homeostatic mechanisms that maintain signaling balance. For example, receptor tyrosine kinases (RTKs) activate both the PI3K/AKT and RAS/ERK pathways that co-regulate pro-survival activity, at least in part, through converging on mTOR signaling32,33. As described in Figure 2, inhibition of either PI3K/AKT or the RAS/ERK pathway can lead to compensatory activation of the other pathway with subsequent maintenance of mTOR signaling and cell survival34. RTK signaling can be upregulated transcriptionally35, or post-translationally by decreased internalization and degradation36, decreased proteolytic shedding of receptors37, formation of novel RTK complexes38 and increased production of receptor ligands39,40. Together these processes engage a suite of mechanisms that could be targeted to interfere with bypass mechanisms and thus increase utility of signal transduction inhibitors.
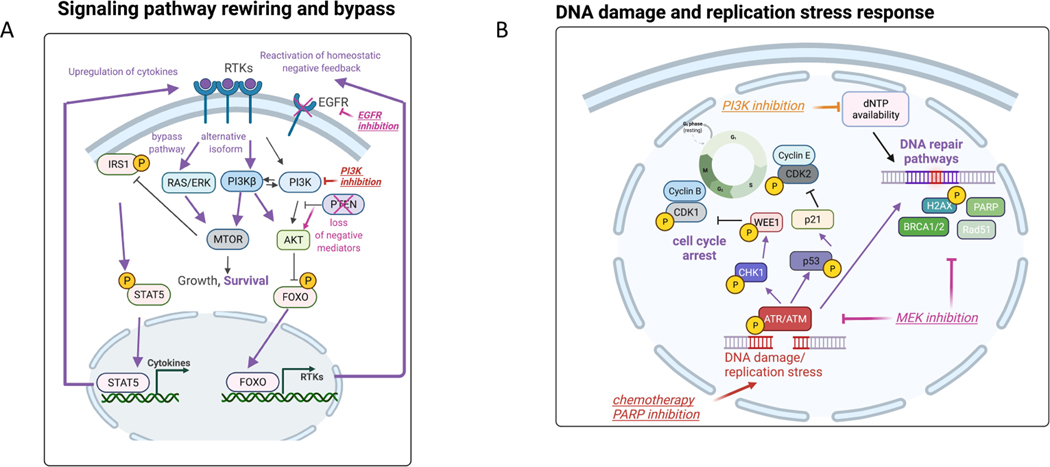
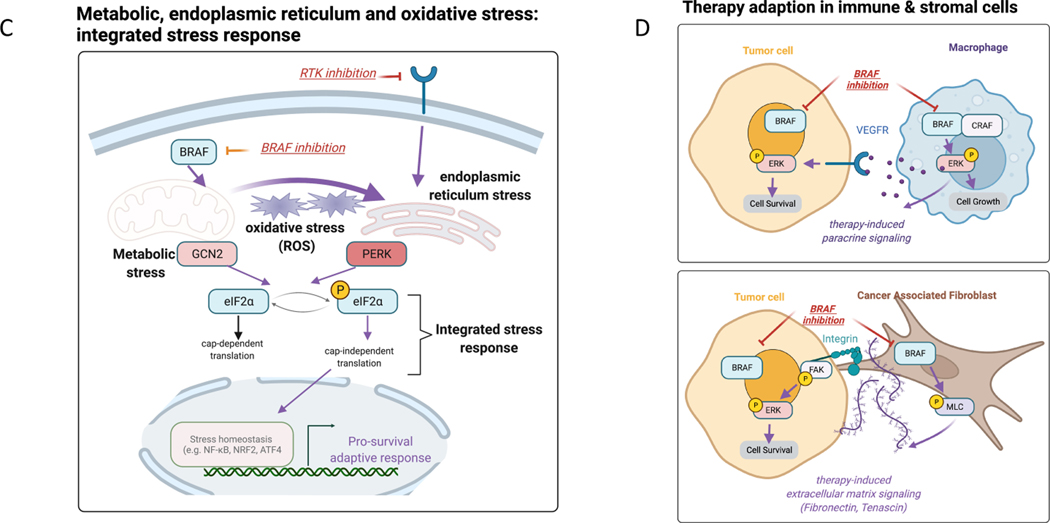
Figure 2: Adaptive response programs in tumor cells and in the tumor ecosystem.
(A) Examples of adaptive responses that bypass inhibition of the PI3K/mTOR pathway: 1) Transcriptional upregulation of RTKs via FOXO transcription factors, 2) secretion of autocrine growth factors via release of the mTOR-driven IRS1 negative feedback loop and 3) utilization of a non-targeted isoform of PI3K. We also highlight another example of signaling rewiring following EGFR inhibition due to downregulation of negative mediators in the PI3K pathway such as PTEN. Purple bolded arrows indicate adaptive responses following each targeted therapy (e.g. EGFRi, PI3Ki, PARPi, MEKi). (B) Engagement of DNA repair pathways to mitigate DNA damage and replication stress. DNA damaging anti-cancer therapies (examples of chemotherapy and PARP inhibition) induce activation of DNA integrity sensors ATM/ATR that induce cell cycle arrest and active DNA repair pathways. PI3K inhibition promotes DNA damage by depletion of nucleoside availability that is required for DNA repair. MEK inhibition represents another example that decreases the levels of DNA repair pathway members resulting in elevated DNA damage. (C) Stress response pathways feed into kinases that phosphorylate eIF2α to induce cap-independent translation of transcription factors that regulate stress homeostasis (integrated stress response). For example, BRAF inhibition mediates metabolic stress via upregulating the upstream kinase GCN2, while RTK inhibition results in PERK upregulation and endoplasmic reticulum (ER) stress. Targeted therapies (e.g. BRAFi) can also result in unbalanced generation of reactive oxygen species (ROS) that via the same eIF2α kinases engage ROS-mitigation programs that are regulated by transcription factors such as NRF2 and NFkB. (D) Tumor-immune paracrine signaling following BRAF inhibition that mediates immunoevasion (proliferation of pro-tumor M2 macrophages) and reactivation of ERK in tumor cells via macrophage-derived VEGF. BRAF inhibition has also been shown to reactivate focal adhesion kinase-driven ERK in melanoma cells via cancer associated fibroblast-mediated extracellular matrix remodeling. Purple bolded arrows indicate adaptive responses resulting from cell-cell communication.
Section 1.2: Adaptive responses that ameliorate cell death pathway activity
Dysregulated cell death programs including apoptosis, ferroptosis, pyroptosis, and necroptosis play critical roles in therapeutic resistance41. Many cellular compensatory mechanisms and stress mitigation pathways shift the balance of cell death pathways towards cell survival. In the case of apoptosis for example, these programs involve induction of anti-apoptotic proteins (e.g. BCL2 family proteins such as BCL2, BCLxL, MCL1) and inactivation or downregulation of pro-apoptotic proteins (e.g. p53, BH3-only BCL2 family proteins). Inhibitors of PI3K and MEK and other cancer therapies can induce stabilization and transcription of BIM and other BH3 family proteins42–45, thus promoting a pro-apoptotic state. However, this is often balanced by upregulation of anti-apoptotic BCL2-family proteins (Table 1). These BCL2-family proteins can be induced by multiple programs at the level of transcription, alternate IRES-dependent translational46,47, or post-translational stabilization48.
Section 1.3: Adaptive responses induced by DNA damage and replication stress
Under conditions where DNA integrity is compromised (e.g. mutation, metabolic stress or environmental toxins), a suite of cell cycle arrest and DNA repair pathways are activated to mitigate effects of the DNA damage49–51. In the presence of DNA damage in normal cells, DNA integrity sensors such as ATM or ATR proteins are recruited to sites of DNA damage where they activate p53 and induce DNA damage repair pathways and cell cycle arrest52 (Figure 2B). p53 is maintained at low levels in unstressed cells by the E3 ubiquitin ligase MDM2 that targets p53 for degradation. In normal cells, multiple stress signals, including oncogenic stress and DNA damage, stabilize p53 with subsequent activation of stress mitigation pathways. Thus, p53 serves as a major cellular stress sensor. Loss or mutation of p53, the most common genomic aberration in cancer, compromises the G1 checkpoint allowing stressed or metabolically compromised cells with decreased dNTPs available for accurate DNA synthesis and repair to enter S phase. Loss of p53 function also allows accumulation of DNA damage by suppressing DNA damaged-induced apoptotic response53. Furthermore, p53 targets multiple genes involved in mismatch repair, nucleotide-excision repair and base-excision repair resulting in dysfunctional DNA repair53. Taken together, loss or mutation of p53 compromises multiple cellular stress mitigation processes allowing accumulation of DNA damage.
Cancer therapies can directly induce DNA damage resulting in activation of the DNA damage response (DDR) pushing tumor cells with high levels of DNA damage over the threshold into cell death. Further a number of chemotherapies decrease the availability of dNTPs required to repair DNA damage, such as gemcitabine that targets RRM2 (Table 1). Several targeted therapies decrease the levels of members of key DNA damage pathways54,55 such as MEK inhibitors compromising homologous recombination in RAS mutant tumors. Tumor initiation and progression is also associated with replication stress (RS) which is defined as failure to protect the replication fork in S phase resulting in replication fork stalling or collapse with subsequent accumulation of DNA damage (Figure 2B). Activation of oncogenes and/or loss of tumor suppressors can elevate RS through: 1) premature S phase entry and a resultant dNTP shortage, 2) stalled replication forks due to replication and transcriptional imbalance, and 3) replication fork collapse56. Interestingly, oncogenes that induce RS and or DNA damage frequently engage stress mitigation pathways56; for example, KRAS mutations can increase HR competence54,57 and MYC activation can resolve transcription-replication conflicts, upregulate dNTP synthesis and increase HR competence58,59.
A number of chemotherapy and targeted agents increase RS by depleting dNTPS or by inducing DNA damage including crosslinks and adducts that result in stalled polymerase60–63 (Table 1, Figure 2B). Drugs that induce replication fork collapse and subsequent double strand DNA breaks (DSBs), such as poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi)64, lead to activation of the S and G2 cell cycle DNA damage checkpoints to allow resolution of the stress (S phase checkpoint) or repair of the damage (G2 phase checkpoint) to occur through HR and other repair pathways prior to entry into M phase. Unresolved RS results in phospho-RPA deletion and further accumulation of DSBs as well as dsDNA in micronuclei and the cytosol65. Cytosolic DNA can lead to activation of the STING pathway, with resultant interferon induction and engagement of the innate and adaptive immune system66–69.
Section 1.4: Adaptive responses and stress mitigation pathways involving metabolic, endoplasmic reticulum and oxidative stress
Metabolic stress:
In normal cells, metabolic plasticity allows cells to quickly adapt to energy and metabolite requirements during conditions where nutrients are limiting. While oxidative phosphorylation (OXPHOS) is the dominant source of energy for slow-growing or stem cells, glycolysis is the preferred energy source for highly proliferative cells. The ability to switch between glycolysis and OXPHOS is controlled by master regulators such as AMP-activated protein kinase (AMPK) and HIF-1, which act as perturbation sensors70,71. Many different cancer therapies induce metabolic stress including PI3K pathway inhibitors that decrease glucose and amino acid uptake72 or BRAF inhibitors that impact glycolysis73,74 (Table 1). For example, BRAF mutant melanoma cells that are reliant on glycolysis are more sensitive to the BRAF inhibitor vemurafemib74, while resistant cells display upregulation of the mitochondrial biogenesis coactivator PGC1a, via the melanocyte master regulator MITF, leading to increased mitochondrial respiration, adaptive resistance to therapy, and sensitivity to OXPHOS inhibition73. Cellular adaptation to BRAF inhibition in melanoma cells depends on the general control nonderepressible 2 (GCN2) kinase, an eIF2α kinase that senses amino acid levels75 (Figure 2C).
Oxidative stress:
Tumor cells often exhibit high oxidative stress as a consequence of unbalanced generation and elimination of reactive oxygen species (ROS) and reactive nitrogen species (RNS) resulting from internal and external metabolic stresses as well as from reduced cellular anti-oxidant levels76–78. Further, aberrant tumor cell metabolism can elevate ROS and RNS77. Most chemotherapies (anthracyclines, platinum coordination complexes, topoisomerase inhibitors, etc.) and many targeted therapies (Figure 2C) that alter nutrient uptake and oxidative phosphorylation (e.g. inhibitors of EGFR, BRAF, HER2, and others) increase ROS/RNS through alterations in mitochondrial electron transport and reduction in anti-oxidant systems like glutathione, peroxidases, superoxide dismutases, and others79 (Table 1). The two broadest-acting ROS-mitigation programs are regulated by NRF2 and NFκB80; these transcription factors induce expression of genes that serve as ROS scavengers and prevent apoptosis associated with high ROS81.
Endoplasmic reticulum (ER) stress:
ER stress response is triggered by various stressors such as hypoxia, metabolic stress or misfolded proteins82,83. Although a strong activation of the ER stress response can lead to cell death, a moderate level of activity can be protective. Thus ER stress response can support both survival and therapy resistance of tumor cells. For example, RTK-targeted therapy in endometrial and renal cancers activates a PERK-driven ER stress response program, resulting in activation of autophagy and NF-κB signaling stress mitigation pathways that promote cell survival84,85 (Table 1, Figure 2C).
Section 1.5: Multiple stress converge on the “integrated stress response” program.
Many of the stress response pathways described above feed into one more kinases (e.g. PERK, GCN2) that phosphorylate eIF2α. Phosphorylation of eIF2α promotes formation of an eIF2-eIF2B inhibitory complex that suppresses global cap-dependent translation, while at the same time promoting cap-independent translation of a subset of mRNAs that regulate homeostasis86,87. Together, these eIF2α kinases integrate signals from many different stresses to induce a common set of stress mitigation programs referred to as the ‘Integrated Stress Response’ (Figure 2C). Therapy-induced stress can overwhelm the integrated cell stress response in cancer cells, thus resulting in cell death88. Further, the switch from cap-dependent to cap-independent protein translation including the anti-apoptotic factor MCL1 can expose novel therapeutic opportunities such as targeting the apoptotic pathway. While this manuscript focusses on a number of independent stress pathways and mitigation process, it is important to acknowledge that they converge, in many cases, on the integrated stress pathway as a final downstream effector.
Section 2: Adaptive responses in tumor stromal cells
Adaptive responses to therapy can also be mediated by tumor-host interactions, where cell types in the tumor ecosystem, produce signals that engage stress mitigation pathways in tumor cells. In addition, therapy-induced alterations in the tumor ecosystem can reprogram stromal cells toward either a pro- or anti-tumor phenotype. Importantly, therapy-induced alterations in tumor-host interactions can reactivate signaling pathways and establish emergent therapeutic opportunities.
Section 2.1 Tumor rewiring during adaptive responses and stress mitigation alters immune contexture
Therapy-induced alterations in the tumor ecosystem can create additional vulnerabilities that can be exploited to enhance therapy efficacy. In order to mediate immunoevasion, tumors produce soluble factors that program both innate and adaptive immune cells in the tumor microenvironment to allow tumor progression89. For example, in melanoma BRAF inhibition can recruit and reprogram macrophages into a protumorigenic M2 phenotype and increase macrophage-derived secretion of VEGF that reactivates ERK in melanoma cells leading to therapeutic resistance90 (Figure 2D). Further, targeted therapy designed to inhibit tumor cell survival can also directly impact immune cell function. The multi-RTK inhibitor sorafenib impairs dendritic cell function and induction of antigen-specific T-cells91, while MEK inhibition can increase anti-tumor activity of CD8+ T cells92. Thus, therapeutic targeting of tumor cells can impact immune population composition, exacerbate tumor supportive immune mechanisms through multiple different mechanisms93, or alternatively can induce an anti-tumor immune context. However, it is clear that the effects of targeting signaling pathways on immune cell phenotype and functional outcomes is nuanced and requires further exploration in model systems and clinical trials to develop approaches that shift the multiple types of immune cells present from an immune evasive microenvironment to a tumor suppressive immune contexture94. Hence, further investigation of therapy-induced adaptation in the complete tumor ecosystem of samples from patients on therapy95,96 will be necessary to inform effective clinical translation.
Section 2.2: Interactions with cancer associated fibroblasts and extracellular matrix
Drug resistance can be sustained by a plethora of growth factors and cytokines released by the neighboring tumor microenvironment (TME), composed by adipocytes, cancer-associated fibroblasts (CAFs), immune and endothelial cells97. Compared to normal fibroblasts, CAFs release elevated levels of pro-inflammatory cytokines and growth factors98. Each of these TME cell types can be influenced by juxtacrine and paracrine interactions with tumor cells. For example, CAFs associated with different tumor types undergo distinct state changes99. Secreted factors from CAFs can in turn influence tumor cells states, including reactivation of signaling pathways that limit the effectiveness of kinase inhibitors in multiple solid tumors100–103 and these pathways can be modulated by both cytotoxic and targeted therapies100,104,105. Extracellular matrix (ECM) remodeling driven by CAFs treated with BRAF inhibitors106 promotes cell survival (Figure 2D). Furthermore, cell-ECM interactions can mediate resistance to PI3K/mTOR and HER2-targeted therapy by adaptive responses involving upregulation of anti-apoptotic Bcl-2 family signaling in ovarian and breast cancer46,107,108. For a comprehensive discussion of effects of cancer therapies on the diverse cell types in the tumor ecosystem, we refer the reader to other review articles16,109.
Section 3: Strategies to exploit tumor cell vulnerabilities associated with adaptive responses to therapies
Tumor intrinsic therapeutic stress and stress mitigation processes engaged by the tumor ecosystem provide three independent therapeutic strategies (Figure 1B) to enhance efficacy of cancer therapies a) block the stress mitigation pathways that allow tumor cells to survive ongoing stress to induce death, b) accentuate existing stress and thus overwhelm mitigation strategies resulting in stress-induced catastrophe and c) exploit emergent vulnerabilities in the tumor ecosystem. In this section, we provide examples of each of these strategies (Figure 3 and Table 2), describe the challenges in deploying these strategies due to tumor heterogeneity (intratumoral and interlesional) (Box 1).
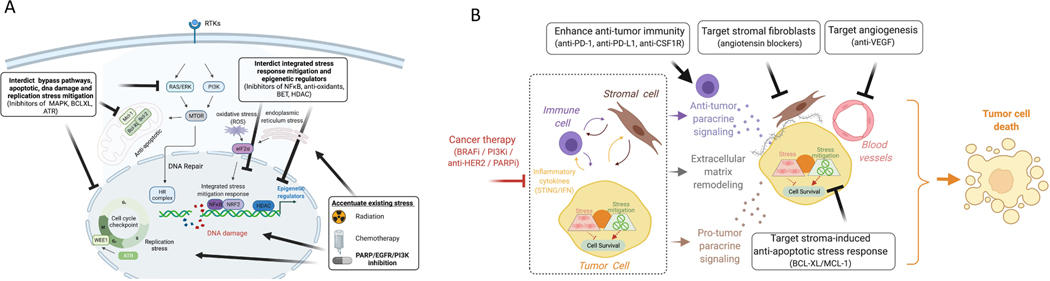
Figure 3. Strategies to induce tumor cell death by exploiting stress adaptation, accentuating existing stress and targeting emergent vulnerabilities in the tumor ecosystem.
(A) Example of therapeutic opportunities emerging from therapy-induced stress. PARP inhibitors induce DNA damage and replication stress that results in cell cycle arrest and engagement of DNA repair pathways. This diagram show strategies involving combination therapies to enhance the efficacy of PARP inhibition that increase stress via radiation or chemotherapy. In addition, there are multiple approaches to interdict the stress mitigation process via 1) blocking anti-apoptotic survival proteins such as BCL-XL, 2) inhibiting upstream mediators of DNA repair (RAS/ERK inhibition) or cell cycle checkpoints (ATR), 3) blocking epigenetic reprograming (HDAC/BET) and 4) stress mitigation programs (e.g. anti-oxidant NRF2, NFkB). (B) Targeting emergent opportunities in the tumor ecosystem represents another approach to develop rational combination therapies. Tumor cells in therapy-resistant tumor ecosystems can be targeted by 1) inhibiting immune checkpoints to augment anti-tumor immunity that is mediated via tumor-immune paracrine signaling, 2) blocking therapy-induced extracellular matrix remodeling that enhances recruitment of cytotoxic immune cells, 3) targeting anti-apoptotic signaling that is induced via stromal-derived cytokines and extracellular matrix remodeling, and 4) blocking angiogenesis to normalize blood vessels and enhance immune cell recruitment.
Table 2 – Therapeutic opportunities created by adaptive responses to cancer therapies.
Combination therapies are organized based on therapeutic approach and the primary therapy that induced the adaptive response.
| Therapeutic strategy | Primary therapy | Combination therapy partner | Cancer Type | Ref |
|---|---|---|---|---|
| Exploit tumor rewiring: block bypass pathway reactivation | ||||
| Blocking signaling reactivation via upstream target inhibition | MTORi | IGF-1Ri | breast | 111 * |
| Blocking multiple members of the same kinase family | PI3Kαi | PI3Kβi | breast | 110 |
| Blocking pathway activation via downstream target inhibition | BRAFi | FAKi | melanoma | 106, 219 * |
| Blocking pathway activation via downstream target inhibition | MTORi | MAPKi | breast | 220 * |
| Interdict stress mitigation programs | ||||
| Targeting DNA damage repair checkpoints | PARPi | ATRi or WEE1i | ovarian | 126 * |
| Blocking homologous recombination repair competency | PARPi | MEKi | ovarian | 54,130 |
| Targeting anti-oxidant to prevent adaptation to elevated oxidative stress | PARPi | TrxR1i | prostate | 142 |
| Inhibiting autophagy to prevent adaption to endoplasmic reticulum stress | multi-RTKi | chloroquine | endometrial | 84 |
| Blocking metabolic reprograming | CDK4/6i | GLS-i | breast& colorectal | 138 |
| Inhibiting anti-apoptotic proteins to target multiple stress mitigation pathways | MAPKi, PI3Ki, HER2i | BCL-XLi, MCL-1i | multiple | 46,116–120,221 * |
| Preventing upregulation of pathway reactivation by targeting transcriptional reprograming | HER2i | BETi | breast | 123 |
| Blocking prosurvival reprogramming by targeting transcriptional activity | PI3Ki | anti-estrogen Receptor | breast | 222 * |
| Increase therapy-induced stress | ||||
| Overloading DNA damage repair pathways via chemotherapy-induced stalled replication forks | gemcitabine | PARPi | lung | 148 |
| Overloading DNA damage repair pathways via irradiation | radiotherapy | PARPi | Ewing sarcoma | 149 |
| Replication stress increased due to decreased nucleotide synthesis | PI3Ki | PARPi | breast cancer | 61 |
| Synergistic cell death via increased reactive oxygen species generation | EGFRi | HDACi | lung cancer | 223 |
| Elevated endoplasmic reticulum stress overwhelms unfolded protein response pathway | EGFRi | PDK1i | lung cancer | 151 |
| Exploit emergent vulnerabilities in the tumor ecosystem | ||||
| Reactivation of CD8+ T cells that were recruited via PARPi-induced STING upregulation | PARPi | anti-PD-L1 | multiple | 69, 95 * |
| Targeting macrophages that reactivate ERK in melanoma cells via VEGF secretion | BRAFi | anti-CSF1R | melanoma | 90 * |
| Enhancing recruitment of cytotoxic T cells by normalized blood vessels | anti-PD-1, anti-CTLA4 | AGTR1i, AGTR2i | breast cancer | 158, 224 * |
References with clinical evidence of adaptive responses to the primary therapy.
Box 1: Heterogeneity of adaptive responses and mitigation pathways:
Cancer cells can adopt a broad range of adaptive responses to survive therapeutic stress. For example, the adaptive rewiring of RTK activity in response to the EGFR/HER2 inhibitor lapatinib was heterogenous across a suite of HER2-positive breast cancer cell lines123. Interestingly, although adaptive responses are heterogeneous across cancer cells, convergence into downstream pathways have been observed. For example, although different RTKs have been shown to be induced across tumor cell lines treated with HER2 inhibitors, bromodomain inhibition blocked the induction of the majority of kinases, suggesting that this epigenetic regulator mediated a common mitigation mechanism123. Further, profiling the proteomic response of ovarian cancer patient derived xenografts treated with PI3K/AKT/mTOR and MAPK pathway inhibitors also revealed heterogeneous patterns of induced RTK expression, but a common coordinated induction of anti-apoptotic proteins116,225. The heterogeneity of adaptive responses engaged by cancer therapies engenders a critical question relating to the extent to which specific stress mitigation pathways adopted by a tumor to bypass therapy stress are driven by underlying genomic aberrations or epigenetic cell states. As a corollary question, can cells be steered with therapy to a particular state with limited mitigation pathways being available to escape therapeutic stress? For example, BRD4 inhibitors have been shown to block expression of multiple kinases precluding the upregulation of RTK activation as a mitigation strategy potentially representing a more therapeutically tractable state226,227.
Importantly, while the impact of intratumoral and interlesion genetic heterogeneity on drug response has been well documented in various cancer types, there is currently little information about the impact of that heterogeneity on adaptive responses to anticancer therapy228. Many genomic alterations converge into a limited set of functional events and protein networks229. Thus, it is possible that a tumor underlying molecular features dictate the possible adaptive resistance mechanisms and that increased tumoral heterogeneity has limited effect on the availability of those adaptive responses. Clearly, further investigation with single-cell analyses will be needed in order to determine the heterogeneity of adaptive responses across cells from a single tumor and across multiple metastatic sites. It will also be important to better determine the impact of intratumoral and interlesion genetic heterogeneity on the variety of available stress mitigation pathways for a given tumor.
Our team has shown that adaptive responses to PARP inhibition can vary greatly across different cell lines as well as patient tumor samples. Adaptive responses included the induction of the DNA damage checkpoint and the PI3K-AKT-mTOR and RAS-MAPK viability pathways126. Importantly, differences were observed in the directionality (e.g. more or less DNA damage), magnitude (e.g. extent of RAS/MAPK activation) and selection of mitigation pathways both within a cell line panel and across patient samples126. The lack of uniformity in mitigation strategies engaged by different tumors highlights the challenge of developing therapies designed to interdict adaptive responses to targeted therapies. Despite the heterogeneity, there appear to be a limited number of programs able to mitigate the therapeutic stress induced by PARPi suggesting that if the mitigation strategy engaged by a particular tumor could be predicted or measured following initiation of therapy, it would be possible to select drug combinations likely to provide patient-specific benefit. Interestingly, although high-grade ovarian cancer tumors have high inter-lesion heterogeneity230,231, our study revealed conserved adaptive responses in different lesions within the same patient126, suggesting that targeting patient-specific mitigation strategies could be effective across different metastatic sites.
3.1. Strategies to block stress mitigation pathways that allow tumor cells to survive ongoing stress
Blocking bypass of signal transduction pathway activation:
Cell survival resulting from adaptive signaling rewiring can be targeted by blocking bypass pathways or inhibiting redundant kinase members in the same family (Table 2, Figure 3A). For example, blocking multiple PI3K family members using PI3K-α/β inhibitors enhances antitumor efficacy in combination with multiple therapies in preclinical studies110. Targeting bypass pathways involving the PI3K/AKT and RAS/MAPK signaling axes can be accomplished by inhibiting receptor tyrosine kinases that are activated by negative feedback loops. For example, reactivation of AKT in breast cancer cells treated with mTOR inhibitors can be blocked using IGF-1R inhibitors111. To combat the heterogeneity and redundancy of receptor tyrosine kinase adaptive responses, pathway reactivation can be targeted by inhibiting downstream kinases. Blocking focal adhesion kinase which is downstream of multiple RTKs can effectively induce cancer cell apoptosis following treatment with HER2-targeted agents in breast cancer112 and with BRAF inhibitors in melanoma106. RAS/MAPK therapy-induced activation of the PI3K/AKT/mTOR stress mitigation pathway in pancreatic cancer113 and melanoma114 has been prevented using AKT and mTOR inhibitors. These examples provide evidence that coordinated inhibition of a target along with the adaptive responses used as a mitigation strategy can prevent bypass and convert cell survival mediated by adaptive responses to cell death. Although this predicts that coordinate targeting of the PI3K/AKT and RAS/ERK pathways would be highly effective, this combination therapy is associated with marked toxicity and is not tolerated in patients115.
Blocking apoptotic stress mitigation:
Multiple targeted therapies (RTK, PI3K/mTOR, PARP and RAS/MAPKi inhibitors) have been shown to result in coordinate upregulation of pro-apoptotic and anti-apoptotic proteins that prime tumors cells for induction of apoptosis by BCL2-family inhibitors. Specifically, blocking BCL-2, BCL-XL or MCL-1 activity has been shown to synergize with RTK, PI3K/AKT and RAS/MAPK pathway blockade in multiple tumor lineages116–120 (Figure 3A, Table 2). Thus, targeting anti-programmed cell death proteins that are downstream of stress mitigation pathways represents an attractive therapeutic approach to increase the efficacy of many different targeted and chemotherapies. Notably, since multiple stress mitigation programs lead to elevation of anti-apoptotic proteins, targeting these proteins represents a potentially more broadly effective therapeutic approach than others that are specifically associated with a single stress mitigation program.
Blocking epigenetic factors:
Targeting epigenetic regulators of cellular programs also represents a candidate approach to inhibit multiple adaptive responses. In melanoma and other cancer types, histone marks alterations, such as the loss of H3K4me3, H3K27me3 and gain of H3K9me3 in response to therapeutic stress has been linked to the development of multidrug resistance121,122. Importantly, silencing methyltransferases can reverse the resistant phenotype. In another study, a BET (BRD4) inhibitor (JQ1) blocked the adaptive stress mitigation response involving upregulation of multiple receptor and non-receptor kinases following HER2 targeted-therapy in breast cancer cells123 (Figure 3A, Table 2). In addition, novel approaches to inhibit formation of active enhancer complexes with a CDK7/12 inhibitor blocked the transcriptional activation of survival programs in response to multiple RTK inhibitors and resulted in enhanced tumor cell killing in vitro and tumor regression in vivo124. Thus, combination therapies that target a key tumor driver while concurrently targeting transcriptional regulators that mediate stress mitigation pathways can increase the activity of a broad suite of drugs and block multiple stress mitigation pathways. Although inhibition of epigenetic regulators that mediate stress mitigation programs engaged downstream of a wide range drugs can reverse adaptive resistance; these combinations have been plagued by toxicity in solid tumors125.
Blocking stress responses associated with DNA damage or RS:
As described above, therapy-induced DNA damage and RS stress mitigation pathways serve as targets to enhance drug efficacy. For example, because PARPi engage the S and G2 DNA damage stress mitigation checkpoints, PARPi combined with inhibition of the ATR/CHK1/WEE1 cascade, which is responsible for activation of the S phase and G2 damage checkpoints, induces synergistic cell death through mitotic catastrophe, and tumor regression in vivo in ovarian and lung cancer models126–128. A number of studies, albeit controversial, suggest that inhibitors of ATR/CHK1/WEE1 cascade are more active in cells that lack p53 due to an increased dependence on the S phase and G2 damage checkpoints129. In addition to engaging DNA damage checkpoints, PARP inhibition can increase homologous recombination (HR) repair competency via RAS/MAPK pathway activation that can be interdicted using MEK inhibitors to sensitize cells to the effects of PARPi130 (Table 2, Figure 3A). Combinations of PARPi with ATR/CHK1/WEE1 or MEK inhibitors have been translated to clinical trials with promising early activity. Importantly, the optimal combination partner for effective PARPi therapy depends on which DNA repair pathways are activated in each tumor. For example, ATR inhibition is most effective in ATM-deficient models127, gemcitabine in cells with premature cell cycle entry and limited dNTP stores 131, and WEE1 inhibition in tumors with high endogenous RS132,133.
Blocking metabolic stress mitigation programs:
Several different approaches to target metabolic adaptations to therapies have also been described, e.g. increasing therapy-induced metabolic stress by coordinate inhibition of the pentose phosphate shunt and glutaminolysis to limit generation of anti-oxidant mediators and render cells sensitive to reactive oxygen species (ROS) generated by OXPHOS134,135. The effects of decreasing or increasing ROS, as with many other targets, is likely to be complex as ROS not only have effects on tumor viability but can influence metastatic potential as well as immune activity136,137. In addition, treatment of breast and colorectal tumors with CDK4/6 inhibitors results in metabolic reprogramming with a shift to utilization of glutamate as an energy source that can be exploited by targeting glutaminase138 (Table 2). While it is clear that adaptation to metabolic stress is a mitigation strategy utilized by both tumor and normal cells, currently there are no facile clinically applicable approaches to identify and target the mitigation processes engaged by the different cell populations in individual tumors.
Blocking oxidative stress mitigation programs:
Development of effective targeted inhibitors of anti-oxidant stress mitigation programs presents challenges in specificity, pharmacodynamics, efficacy and safety139,140. However, inhibition of targets of NRF2 and NFκB such thioredoxin or glutathione generating enzymes can enhance drug sensitivity141. For example, the elevated oxidative stress induced by PARPi that contributes to the induction of DNA damage and cell death142, can be exploited by inhibiting the anti-oxidant enzyme thioredoxin reductase to induce tumor regression in prostate cancer xenografts128. Further work is required to develop optimal anti-oxidant dosing strategies and delivery to avoid toxicities and develop a therapeutic index in cancer patients.
Section 3.2: Strategies to increase ongoing and therapy-induced stress to induce cellular catastrophe
As indicated above, sustained therapeutic stress in tumor cells results in the engagement of stress mitigation programs. If the stress can be increased to a level that overwhelms the stress mitigation programs, this ultimately results in tumor cell death. Given that cancer cells, as opposed to normal cells, are subject to ongoing stress, combination therapies that exploit this differential stress level could provide an adequate therapeutic index. Previous studies have shown that normal cells compared to tumor cells exhibit lower baseline levels of stress143,144. To explain the increased sensitivity of tumor cells compared to normal cells to a number of stress-inducing therapies, it has been proposed that stress-induced catastrophe when cell pass a common stress threshold145,146. This is particularly evident in cases such as RS or apoptosis where catastrophe or cell death occurs above a stress threshold65,147. Several approaches have been used to increase RS or to target pathways that mitigate the effects of RS60,62. Addition of gemcitabine or PI3K inhibitors to PARPi can significantly increase RS by depleting dNTPs thus overloading RS mitigation pathways resulting in accumulation of DNA damage, S phase and mitotic catastrophe and tumor growth inhibition61,148. Overloading the DDR pathways and the DNA damage checkpoint mitigation strategy can be induced by combining PARPi with radiotherapy (Figure 3A) resulting in increased double strand breaks, apoptosis and slower tumor growth in Ewing’s sarcoma models149. We have demonstrated that an equal increase in ongoing RS in tumor cells induced by sequential therapy with PARP and WEE1 inhibitors induces death of tumor cells while concurrently sparing normal cells and mitigation of toxicity132. The concept that ongoing RS in tumor cells with low RS in normal cells could result in a therapeutic index is being tested in an ongoing clinical trial (NCT0419773).
Exacerbating the metabolic, ER and oxidative stress pathways described above can also overwhelm mitigation pathways and result in cell death through apoptosis or other cell death mechanisms such as necrosis, ferroptosis and autophagic cell death150. For example, EGFR inhibition has also been shown to elevate ER stress (Figure 3A) and in combination with PDK1 inhibitors increase ER stress to a level that overwhelmed the mitigation pathway and resulted in cell death and improved survival in lung cancer xenografts151. Thus, combination therapies that effectively increase therapy-induced stress levels in tumor cells provide a promising strategy to block cell survival by inducing stress overload that can overwhelm stress mitigation processes and this approach could lead to a therapeutic index.
Section 3.3: Targeting stroma cell adaptive responses
Immune cell responses:
Both the pro and anti-tumor immune effects induced by therapeutic targeting provide novel opportunities for intervention. For example, in melanoma targeting CSF1R on M2 macrophages reduced their recruitment and reduced tumor growth rates in xenografts90. PARPi can induce both anti- and protumorigenic macrophages through metabolic reprograming in BRCA1-deficient breast cancer syngeneic models152. The protumorigenic effects mediated by PARPi-induced CSF1 production by tumor cells can be reversed by targeting CSF1R on macrophages.
Conversely, activation of the STING pathway by PARPi in lung, colorectal, breast and ovarian tumors leads to production of interferon that can activate an innate immune response that, in turn, induces mobilization of CD4+ and CD8+ T cells68,153 (Figure 3B, Table 2). Treatment with immune checkpoint inhibitors (ICI) against PD-1 or PD-L1 capitalized on this therapy-induced alteration in the immune tumor ecosystem and induced complete regression and durable responses in both BRCA-proficient and BRCA-deficient tumor models, highlighting the critical role of therapy-induced immune cell recruitment for therapeutic efficacy154. Indeed, multiple PARPi and ICI combination trials are underway95,155. Oncogenic activation can induce an immunosuppressive microenvironment with targeting of oncogene signaling in tumor cells both demonstrating tumorintrinsic activity and altering the tumor microenvironment. For example, mutant KRAS is associated with the production of multiple cytokines that recruit myeloid derived suppressor cells (MDSCs). The recruitment of MDSCs can be reversed by MEK inhibitors with the inhibition of the tumor intrinsic effects increasing tumor cell sensitivity to ICI156. Thus, it is possible to capitalize on new vulnerabilities in the tumor ecosystem resulting from adaptive responses induced by therapeutic targeting that are not directly a consequence of the agent on tumor cells. To translate these findings to the clinic, it is critical to identify recurrent mechanisms of tumor-immune and immune-immune interactions directly in samples from patients on therapy, while considering both the pro- and anti-tumor effects of the tumor microenvironment. Unfortunately, many of our anti-tumor therapies mediate deleterious effects on the immune system (e.g. impaired dendritic cell function by RTK inhibition91). Thus it will be critical to determine the effects of targeting adaptive responses in tumor and the microenvironment on tumor cells as well as on components of the immune system to identify approaches that compromise tumor cell viability while at the same time engaging an effective anti-tumor immune response.
Targeting CAF responses and the vascular barrier:
Tumors adapt to limited nutrient and O2 availability by inducing a neoangiogenic program157. Notably, strategies aimed at targeting CAF-mediated ECM remodeling using angiotensin blockers (Figure 3B) have also been shown to normalize blood vessels leading to enhanced sensitivity to chemotherapy in ovarian cancer xenografts158 and the recruitment of immune cells and improved response to immunotherapy in a syngeneic breast model158, as well as the response to chemotherapy in human pancreatic cancer patients159. Therapies directed towards the vascular barrier (e.g. via the VEGFC/VEGFR3 axis160) represent a stroma-targeted approach (Figure 3B) that has been FDA-approved in combination with cytotoxic chemotherapy or immunotherapy160–162 for a subset of solid tumors (lung, ovary and breast). Taken together, effects of therapies designed to target tumor cells can rewire the extracellular stromal environment providing opportunities to develop an anti-tumor environment.
Section 4. Translation to the clinic
The concept that tumors and the tumor ecosystem are under ongoing stress that is accentuated by therapy underlies the rationale for translating the three proposed approaches (Figure 1B, Table 1) to exploit adaptive responses and resultant mitigation processes to the clinic. Compared to empirical combination therapy design, this concept provides a framework to rationalize choice of drug pairs and biomarkers of therapeutic response. Translating these opportunities to the clinic with subsequent patient benefit requires deploying a complex suite of combination therapies able to: a) target adaptive responses in the clinic by interdicting stress mitigation resulting in cell death, b) increase stress in tumor cells resulting in cellular catastrophe or c) exploit the network rewiring that occurs as cells explore different states and mitigation strategies through combination therapies targeting tumor or stromal cells in the tumor ecosystem. Importantly, the drug combinations able to exploit each of these alternatives are markedly different and need to be deployed in the appropriate manner (potentially with different scheduling and dosing strategies) and in patients most likely to benefit (Figure 4). The combination therapies must secondly exhibit a therapeutic index so that efficacy is displayed at tolerable doses. Indeed, as therapeutic index represents the rate limiting step for utility of any therapy, combination therapies including those targeting adaptive responses are all too often limited by toxicity rather than by lack of efficacy (Box 2)163. The underlying precept we have espoused wherein tumors, in contrast to normal cells, are under chronic stress and that this stress state can be accentuated by therapy provides the potential for a therapeutic index1–3,132.
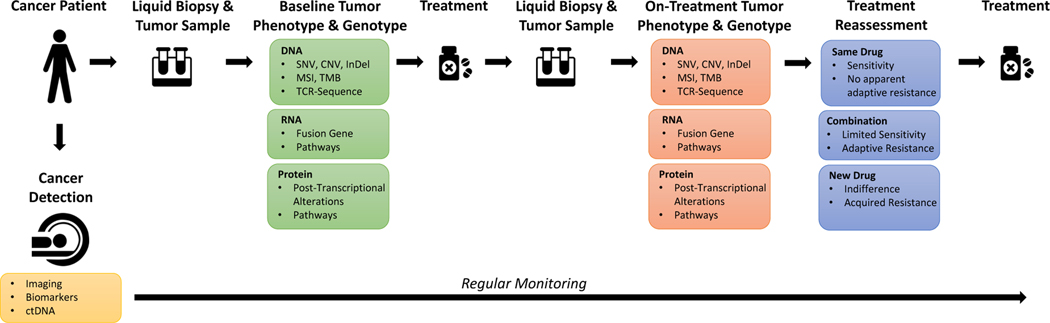
Figure 4. Potential approach to deliver personalized combination therapies in real time.
Liquid biopsy and tumor samples are collected at diagnosis. A suite of CLIA assays are performed to establish the baseline tumor phenotype and genotype of the tumor and select an initial therapy. After a short treatment period, a second liquid biopsy and tumor samples are collected and re-analyzed. The on-treatment tumor phenotype and genotype is established and the therapy is reassessed for one of three scenarios. (1) If sensitivity to therapy is suspected, no changes are made in terms of treatment. (2) If targetable adaptive responses are induced, a second drug is added to generate a patient-specific combination therapy. (3) If the tumor is indifferent to therapy or displays acquired resistance, a new treatment will be selected. Importantly, during the whole process, the progression of the disease is monitored regularly. In case of progression, new samples are acquired and analyzed for therapy reassessment.
Box 2. Limitation of drug combinations targeting stress mitigation pathways.
Based on the current literature, translation of the three proposed approaches (Figure 1B) to exploit adaptive responses and resultant mitigation processes to the clinic has the potential to improve patients care. However, there are still several questions that grants further exploration. For example, it is unclear how different cells in the tumor ecosystem will respond to the therapy. Also, where multiple stress mitigation pathways are activated simultaneously, targeting one might not be sufficient. The heterogeneity in mitigation processes between and within tumors are still poorly described. Other challenges that might be encountered comes from the complexity of some networks and the effects of perturbing them, as well as the impact of the presence of pre-existing resistant clones that could be selected by the proposed approaches.
Although many drug combinations have demonstrated promising results in pre-clinical studies, the additive or synergistic activity often comes at the cost of increased toxicity in patients. This can be associated specifically with acute tumor cell cytotoxicity such as tumor lysis syndromes232 or with more direct effects of the drugs on host organs that are dependent on the targeted pathways. One potential advantage of targeting stress mitigation pathways is that these programs are generally not required for normal tissue homeostasis. Several strategies can be used to improve the therapeutic index. With therapeutic option ‘a’ (Figure 1B), the second drug that inhibits the stress mitigation pathway can be delivered sequentially to reduce toxicity. For option ‘b’ that involves use of drugs that enhance stress, it is important to specifically design combinations that target tumor specific stress programs to avoid systemic toxicities. Therapies that target molecules preferentially expressed in cancer cells, as well as immunotherapies tends to present with a better therapeutic index than cytotoxic agents. For drugs that have similar toxicity profiles (e.g. selumetinib and erlotinib233) dose-escalation to the recommended phase II monotherapy doses is unlikely. Thus, exploring different treatment schedule such as sequential drug administration or periodic drug holiday (intermittent treatment) can be considered132. Finally, delivering or trapping the drugs at the desirable site using nanocarriers or anti-body drug conjugates for example, can improve drug bioavailability selectively in tumor and decrease adverse events affecting normal cells234.
Importantly, a suite of cost-effective CLIA compliant assays that can select combinations for specific patients based either on the pre-therapy state or on the changes that are induced by therapy stress must be deployed. The overarching goal is to develop assays that facilitate selection of therapies targeting adaptive responses and determine when changes in therapies are necessary at a pace that is faster than the tumor can evolve to utilize alternative mitigation pathways. Importantly, the information content in widely used DNA sequence-based analyses is inadequate as the mitigation processes described herein are primarily mediated by non-genomic mechanisms10–16. Assessment of transcriptional changes that mediate adaptive responses or more direct measurement of the protein and metabolic events that mediate the mitigation strategies will be required. The analytic platforms and computational biology pipelines must be able to deliver results to the physician in near real time. The suite of potential approaches to deliver on this opportunity include molecular imaging164–166 or analysis of tissue or liquid biopsies167–169 (Figure 4). Other surrogates such as analyzing immune state in peripheral blood to predict immune contexture in the tumor also hold promise 170,171. Optimally, the CLIA compliant assays should capture the state of tumor cells and the tumor ecosystem across the set of metastatic lesions present in the patient. Liquid biopsies as they integrate information across all of the tumor sites in the body are thus attractive. To fulfill this goal, analysis of circulating tumor cells169, circulating hybrid cells172, RNA173 and miRNA174 and proteins in exosomes175 as they change with therapeutic stress has the potential to alleviate the need for tumor biopsies, allow assessment of multiple time points, make personalized therapy approaches available to a wider set of patients and help to democratize the concept of targeting adaptive responses.
Currently we do not have approved combination therapies that can exploit the broad suite of mitigation strategies engaged by tumors. Fortunately, the repertoire of available drugs is increasing rapidly. Considering both on-target and known off-target activities covers an impressive set of potential targets and is beginning to make targeting adaptive responses and stress mitigation pathways practical. Furthermore, drugging what was previously thought to be undruggable targets by selectively degrading proteins or decreasing protein production through antisense or miRNA therapies is changing the landscape of potentially targetable molecules176–178. Whether CRISPR will prove applicable to cancer therapy and further increase the target repertoire is only beginning to be explored179. Most current targeted therapies target both the wild type and mutated molecule with attendant challenges with toxicity. The emerging ability to target mutant molecules such as KRASG12C while sparing the wild type molecule has resulted in high activity with limited toxicity180,181.
To fully exploit opportunities to target mitigation strategies, novel trial designs with the flexibility to both measure adaptive responses and to rapidly change therapy as the mitigation strategies used by the tumor evolve under therapeutic stress will be needed. Further, as the mitigation strategies engaged by tumors are likely to be patient- or tumor-specific, the trials must allow selection of patient-specific approaches. Novel trials design such as monotherapy run-ins followed by on-therapy biopsies and subsequent combination therapy selection provide the opportunity to identify patient-specific combination therapies designed to exploit adaptive responses182. However, the monotherapy run-in with subsequent combination therapy selection comes with challenges of requiring multiple biopsies (pre- and on-therapy) and also of deploying effective analytics in real time. Longitudinal profiling of adaptive responses coupled with genomic analysis of mutant clone evolution (e.g. estrogen receptor in luminal breast cancer23–25, PI3K in HER2 breast cancer183, MET in lung cancer184 and KRAS in melanoma185) will inform the design of therapies that capitalize on vulnerabilities of the heterogeneous tumor clones with the ultimate goal to prevent the emergence of new resistant clones.
The amount of information that will be available on each patient based on emerging technologies that assess mitigation strategies, the broad array of available drugs particularly considering both on and off target activity, the challenge of predicting toxicity of combination therapies and the need to have up-to-date results from clinical trials to guide patient management are rapidly emerging challenges. Physician support approaches such as highly curated databases linking molecular markers to therapeutic options are sorely needed. Even more importantly, these databases must be easily accessed and readily understood. Artificial intelligence approaches linked to natural language processing have the ability to overcome some of the challenges. However, even the best current approaches require additional validation and curation. Data management and visualization tools will be required for tumor boards to engage the “wisdom of the crowd” that could aid in selecting patient specific combination therapies that effectively take advantage of adaptive responses.
Section 5 Conclusions & Perspectives
Resistance to cancer therapy remains a major challenge. To improve patient outcomes, we need to advance our understanding of the mechanisms that tumor ecosystems employ to survive both ongoing and therapy-induced stress. Single-cell RNA-seq or single-cell proteomics approaches that separately analyze the heterogeneity of tumor, stroma and immune cell states will aid in the discovery of new therapeutic opportunities that can exploit vulnerabilities induced by adaptive responses to therapy at the tumor-stroma interface186–188. Profiling of the complete tumor ecosystem will also advance understanding of tumor microenvironment adaptation to therapeutic stress, given the intense focus on immunotherapy combinations189. The efficacy of combination therapies on tumor cell intrinsic states will need to be balanced with their potentially deleterious effects on the immune system190. Importantly, the effects of stress, stress mitigation pathways and attempts at targeting are likely to be more nuanced than presented herein and will require a much deeper analysis of underlying mechanisms and importantly of the effects of perturbations on tumor, stromal and immune cell states with both temporal and spatial resolution in the clinic and in model systems. Indeed, while this review has focused on using examples to elucidate critical concepts, the effects of therapeutic targeting is likely to be dependent on the particular tumor context, treatment history and also on the complete tumor ecosystem. Further, the complex network of stress mitigation pathways and utilization of different stress mitigation pathways in different tissues and different individuals and the likelihood that multiple different mitigation pathways are initiated in individual tumors will require detailed analysis of the effects of therapies on stress mitigation pathways in patient samples and in particular samples from patients on therapy. The challenges behind obtaining and analyzing samples in depth will require the development of national and international consortia as well as mechanisms to share data in a more efficient manner.
Objective analysis of therapy-induced stress mitigation responses in the clinic will be necessary to develop therapeutic strategies that prevent the emergence of drug resistance. While current analytic approaches are dependent on tumor biopsies, surrogate assays with imaging modalities164–166 or liquid biopsies167–169 to alleviate the cost, technical challenges and morbidity associated with repeat tumor biopsies would be preferable. Molecular imaging-based approaches (e.g. reporters of specific signaling pathways106,191) offer the ability to capture adaptive responses in multiple metastatic sites; an area of critical importance to combat metastatic cancers. Furthermore, longitudinal monitoring will help address the challenge of designing optimal treatment schedules (sequential administration and the order of each inhibitor192 versus simultaneous delivery) to balance on-target toxicity with tumor cell eradication. As the spectrum of clinically-relevant adaptive responses employed in tumors are revealed, high fidelity, spatially resolved tumor models34,193,194 195–197 will be needed to allow longitudinal analysis of tumor cell intrinsic and extrinsic adaptive responses. Importantly, barcoding technologies198 have the potential to allow better characterization of the plasticity of the stress mitigation pathways. Systems-level profiling199,200 of therapy-induced cellular stress homeostasis in diverse tumors will address the open questions of whether different tumor genotypes share a finite suite of adaptive responses and approaches to identify optimal nodes for combination therapy.
Despite the challenges, the opportunity to target adaptive responses and stress mitigation pathways provides a potential to not only directly impact tumor cell survival but also to prevent the emergence of genomic mutation driven resistant clones that are notoriously hard to target while at the same time to engage an effective immune response. Thus, exploiting stress and stress mitigation pathways has the potential to provide major benefits to cancer patients.
Footnotes
Competing interests:
ML and IZ and JB have no COI relating to the topics in this review. GBM: Consultant/Scientific Advisory Board Companies: AstraZeneca, Ellipses Pharma, ImmunoMET, Infinity, Ionis, Lilly, Nanostring, PDX Pharmaceuticals, Signalchem Lifesciences, Tarveda, Turbine, Zentalis Pharmaceuticals Stock/Options/Financial Companies: Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindletop Ventures, Tarveda, Turbine. Licensed Technology: HRD assay to Myriad Genetics, DSP to Nanostring.
REFERENCES
- 1.Galluzzi L, Yamazaki T. & Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol 19, 731–745, doi: 10.1038/s41580-018-0068-0 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Ganesh K. & Massague J. Targeting metastatic cancer. Nat Med 27, 34–44, doi: 10.1038/s41591-020-01195-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Magnen C, Shen MM & Abate-Shen C. Lineage Plasticity in Cancer Progression and Treatment. Annu Rev Cancer Biol 2, 271–289, doi: 10.1146/annurev-cancerbio-030617-050224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chovatiya R. & Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol Cell 54, 281–288, doi: 10.1016/j.molcel.2014.03.030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotamisligil GS & Davis RJ Cell Signaling and Stress Responses. Cold Spring Harb Perspect Biol 8, doi: 10.1101/cshperspect.a006072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo J, Solimini NL & Elledge SJ Principles of cancer therapy: oncogene and nononcogene addiction. Cell 136, 823–837, doi: 10.1016/j.cell.2009.02.024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreuzaler P, Panina Y, Segal J. & Yuneva M. Adapt and conquer: Metabolic flexibility in cancer growth, invasion and evasion. Mol Metab 33, 83–101, doi: 10.1016/j.molmet.2019.08.021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X. & Zhang Z. Understanding the Genetic Mechanisms of Cancer Drug Resistance Using Genomic Approaches. Trends Genet 32, 127–137, doi: 10.1016/j.tig.2015.11.003 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Salgia R. & Kulkarni P. The Genetic/Non-genetic Duality of Drug ‘Resistance’ in Cancer. Trends Cancer 4, 110–118, doi: 10.1016/j.trecan.2018.01.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marine JC, Dawson SJ & Dawson MA Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 20, 743–756, doi: 10.1038/s41568-020-00302-4 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Arozarena I. & Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat Rev Cancer 19, 377–391, doi: 10.1038/s41568-019-0154-4 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Chern YJ & Tai IT Adaptive response of resistant cancer cells to chemotherapy.Cancer Biol Med 17, 842–863, doi: 10.20892/j.issn.2095-3941.2020.0005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fendt SM, Frezza C. & Erez A. Targeting Metabolic Plasticity and Flexibility Dynamics for Cancer Therapy. Cancer Discov 10, 1797–1807, doi: 10.1158/21598290.CD-20-0844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta PB, Pastushenko I, Skibinski A, Blanpain C. & Kuperwasser C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 24, 65–78, doi: 10.1016/j.stem.2018.11.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marzagalli M, Fontana F, Raimondi M. & Limonta P. Cancer Stem Cells-Key Players in Tumor Relapse. Cancers (Basel) 13, doi: 10.3390/cancers13030376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaked Y. The pro-tumorigenic host response to cancer therapies. Nat Rev Cancer 19, 667–685, doi: 10.1038/s41568-019-0209-6 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Holohan C, Van Schaeybroeck S, Longley DB & Johnston PG Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13, 714–726, doi: 10.1038/nrc3599 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Assaraf YG et al. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resist Updat 46, 100645, doi: 10.1016/j.drup.2019.100645 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Bukowski K, Kciuk M. & Kontek R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int J Mol Sci 21, doi: 10.3390/ijms21093233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konieczkowski DJ, Johannessen CM & Garraway LA A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell 33, 801–815, doi: 10.1016/j.ccell.2018.03.025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Wang Z, Ajani JA & Song S. Drug resistance and Cancer stem cells. Cell Commun Signal 19, 19, doi: 10.1186/s12964-020-00627-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward RA et al. Challenges and Opportunities in Cancer Drug Resistance. Chem Rev 121, 3297–3351, doi: 10.1021/acs.chemrev.0c00383 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Mao P. et al. Acquired FGFR and FGF Alterations Confer Resistance to Estrogen Receptor (ER) Targeted Therapy in ER(+) Metastatic Breast Cancer. Clin Cancer Res 26, 5974–5989, doi: 10.1158/1078-0432.CCR-19-3958 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Nayar U. et al. Acquired HER2 mutations in ER(+) metastatic breast cancer confer resistance to estrogen receptor-directed therapies. Nat Genet 51, 207–216, doi: 10.1038/s41588-018-0287-5 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Priedigkeit N. et al. Acquired mutations and transcriptional remodeling in long-term estrogen-deprived locoregional breast cancer recurrences. Breast Cancer Res 23, 1, doi: 10.1186/s13058-020-01379-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priedigkeit N. et al. Intrinsic Subtype Switching and Acquired ERBB2/HER2 Amplifications and Mutations in Breast Cancer Brain Metastases. JAMA Oncol 3, 666671, doi: 10.1001/jamaoncol.2016.5630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakai W. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2mutated cancers. Nature 451, 1116–1120, doi: 10.1038/nature06633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waks AG et al. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann Oncol 31, 590–598, doi: 10.1016/j.annonc.2020.02.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wander SA et al. The Genomic Landscape of Intrinsic and Acquired Resistance to Cyclin-Dependent Kinase 4/6 Inhibitors in Patients with Hormone Receptor-Positive Metastatic Breast Cancer. Cancer Discov 10, 1174–1193, doi: 10.1158/2159-8290.CD19-1390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes D. & Andersson DI Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat Rev Genet 16, 459–471, doi: 10.1038/nrg3922 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Nam AS, Chaligne R. & Landau DA Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet 22, 3–18, doi: 10.1038/s41576-020-0265-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov 2, 311–319, doi: 10.1158/2159-8290.CD-12-0018 (2012). This study shows that AKT inhibition leads to the induction of receptor tyrosine kinases through the relief of a feedback loop, reducing the antitumor activity of the AKT inhibition.
- 33.Fruman DA et al. The PI3K Pathway in Human Disease. Cell 170, 605–635, doi: 10.1016/j.cell.2017.07.029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozengurt E, Soares HP & Sinnet-Smith J. Suppression of feedback loops mediated by PI3K/mTOR induces multiple overactivation of compensatory pathways: an unintended consequence leading to drug resistance. Mol Cancer Ther 13, 2477–2488, doi: 10.1158/1535-7163.MCT-14-0330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarty A, Sanchez V, Kuba MG, Rinehart C. & Arteaga CL Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A 109, 2718–2723, doi: 10.1073/pnas.1018001108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerosa L. et al. Receptor-Driven ERK Pulses Reconfigure MAPK Signaling and Enable Persistence of Drug-Adapted BRAF-Mutant Melanoma Cells. Cell Syst 11, 478–494 e479, doi: 10.1016/j.cels.2020.10.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller MA et al. Reduced Proteolytic Shedding of Receptor Tyrosine Kinases Is a Post-Translational Mechanism of Kinase Inhibitor Resistance. Cancer Discov 6, 382–399, doi: 10.1158/2159-8290.CD-15-0933 (2016). This study demonstrates that extracellular proteomic adaptation can lead to signaling bypass and drug resistance.
- 38.Cooper J. & Giancotti FG Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell 35, 347–367, doi: 10.1016/j.ccell.2019.01.007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Britschgi A. et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 22, 796–811, doi: 10.1016/j.ccr.2012.10.023 (2012). This study shows that JAK2/STAT5 are part of a positive feedback loop that reduces the efficacy of PI3K/mTOR inhibition
- 40. Wang VE et al. Adaptive Resistance to Dual BRAF/MEK Inhibition in BRAF-Driven Tumors through Autocrine FGFR Pathway Activation. Clin Cancer Res 25, 7202–7217, doi: 10.1158/1078-0432.CCR-18-2779 (2019). This research demonstrates that activation of the FGFR pathway represent a bypass mechanism to the dual BRAF/MEK inhibition, leading to adaptive resistance.
- 41.Green DR The Coming Decade of Cell Death Research: Five Riddles. Cell 177, 1094–1107, doi: 10.1016/j.cell.2019.04.024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley R, Balmanno K, Hadfield K, Weston C. & Cook SJ Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 278, 18811–18816, doi: 10.1074/jbc.M301010200 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Harada H. & Grant S. Targeting the regulatory machinery of BIM for cancer therapy. Crit Rev Eukaryot Gene Expr 22, 117–129, doi: 10.1615/critreveukargeneexpr.v22.i2.40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmelzle T. et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci U S A 104, 3787–3792, doi: 10.1073/pnas.0700115104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villunger A. et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038, doi: 10.1126/science.1090072 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Muranen T. et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell 21, 227–239, doi: 10.1016/j.ccr.2011.12.024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherrill KW, Byrd MP, Van Eden ME & Lloyd RE BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem 279, 29066–29074, doi: 10.1074/jbc.M402727200 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Hartman ML, Talar B, Gajos-Michniewicz A. & Czyz M. MCL-1, BCL-XL and MITF Are Diversely Employed in Adaptive Response of Melanoma Cells to Changes in Microenvironment. PLoS One 10, e0128796, doi: 10.1371/journal.pone.0128796 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lans H, Hoeijmakers JHJ, Vermeulen W. & Marteijn JA The DNA damage response to transcription stress. Nat Rev Mol Cell Biol 20, 766–784, doi: 10.1038/s41580-019-0169-4 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Li Y. & Lu X. Regulators in the DNA damage response. Arch Biochem Biophys 594, 18–25, doi: 10.1016/j.abb.2016.02.018 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Petsalaki E. & Zachos G. DNA damage response proteins regulating mitotic cell division: double agents preserving genome stability. FEBS J 287, 1700–1721, doi: 10.1111/febs.15240 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Blackford AN & Jackson SP ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell 66, 801–817, doi: 10.1016/j.molcel.2017.05.015 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Boutelle AM & Attardi LD p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol 31, 298–310, doi: 10.1016/j.tcb.2020.12.011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun C. et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci Transl Med 9, doi: 10.1126/scitranslmed.aal5148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun C. et al. BRD4 Inhibition Is Synthetic Lethal with PARP Inhibitors through the Induction of Homologous Recombination Deficiency. Cancer Cell 33, 401–416 e408, doi: 10.1016/j.ccell.2018.01.019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berti M, Cortez D. & Lopes M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat Rev Mol Cell Biol 21, 633–651, doi: 10.1038/s41580-020-0257-5 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Kalimutho M. et al. Enhanced dependency of KRAS-mutant colorectal cancer cells on RAD51-dependent homologous recombination repair identified from genetic interactions in Saccharomyces cerevisiae. Mol Oncol 11, 470–490, doi: 10.1002/1878-0261.12040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baluapuri A, Wolf E. & Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol 21, 255–267, doi: 10.1038/s41580-020-0215-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi J. et al. MYC status as a determinant of synergistic response to Olaparib and Palbociclib in ovarian cancer. EBioMedicine 43, 225–237, doi: 10.1016/j.ebiom.2019.03.027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baillie KE & Stirling PC Beyond Kinases: Targeting Replication Stress Proteins in Cancer Therapy. Trends Cancer, doi: 10.1016/j.trecan.2020.10.010 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Juvekar A. et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc Natl Acad Sci U S A 113, E4338–4347, doi: 10.1073/pnas.1522223113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin JC et al. Exploiting Replication Stress as a Novel Therapeutic Intervention. Mol Cancer Res 19, 192–206, doi: 10.1158/1541-7786.MCR-20-0651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu H, Swami U, Preet R. & Zhang J. Harnessing DNA Replication Stress for Novel Cancer Therapy. Genes (Basel) 11, doi: 10.3390/genes11090990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rose M, Burgess JT, O’Byrne K, Richard DJ & Bolderson E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front Cell Dev Biol 8, 564601, doi: 10.3389/fcell.2020.564601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toledo L, Neelsen KJ & Lukas J. Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Mol Cell 66, 735–749, doi: 10.1016/j.molcel.2017.05.001 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Coquel F. et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61, doi: 10.1038/s41586-018-0050-1 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Mackenzie KJ et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465, doi: 10.1038/nature23449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shen J. et al. PARPi Triggers the STING-Dependent Immune Response and Enhances the Therapeutic Efficacy of Immune Checkpoint Blockade Independent of BRCAness. Cancer Res 79, 311–319, doi: 10.1158/0008-5472.CAN-18-1003 (2019). This research demonstrates that PARP inhibitors promote the accumulation of cytosolic DNA fragments, activating the cGAS-STING pathway, inducing antitumor immunity.
- 69.Wang Z. et al. Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci Rep 9, 1853, doi: 10.1038/s41598-019-38534-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koyasu S, Kobayashi M, Goto Y, Hiraoka M. & Harada H. Regulatory mechanisms of hypoxia-inducible factor 1 activity: Two decades of knowledge. Cancer Sci 109, 560571, doi: 10.1111/cas.13483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaw RJ Glucose metabolism and cancer. Curr Opin Cell Biol 18, 598–608, doi: 10.1016/j.ceb.2006.10.005 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Park JH, Pyun WY & Park HW Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets. Cells 9, doi: 10.3390/cells9102308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haq R, Fisher DE & Widlund HR Molecular pathways: BRAF induces bioenergetic adaptation by attenuating oxidative phosphorylation. Clin Cancer Res 20, 2257–2263, doi: 10.1158/1078-0432.CCR-13-0898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haq R. et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell 23, 302–315, doi: 10.1016/j.ccr.2013.02.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagasawa I, Kunimasa K, Tsukahara S. & Tomida A. BRAF-mutated cells activate GCN2-mediated integrated stress response as a cytoprotective mechanism in response to vemurafenib. Biochem Biophys Res Commun 482, 1491–1497, doi: 10.1016/j.bbrc.2016.12.062 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Gorrini C, Harris IS & Mak TW Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12, 931–947, doi: 10.1038/nrd4002 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Hayes JD, Dinkova-Kostova AT & Tew KD Oxidative Stress in Cancer. Cancer Cell 38, 167–197, doi: 10.1016/j.ccell.2020.06.001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liou GY & Storz P. Reactive oxygen species in cancer. Free Radic Res 44, 479–496, doi: 10.3109/10715761003667554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang H. et al. The role of cellular reactive oxygen species in cancer chemotherapy. J Exp Clin Cancer Res 37, 266, doi: 10.1186/s13046-018-0909-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rojo de la Vega M, Chapman E. & Zhang DD NRF2 and the Hallmarks of Cancer. Cancer Cell 34, 21–43, doi: 10.1016/j.ccell.2018.03.022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi N. et al. Cancer Cells Co-opt the Neuronal Redox-Sensing Channel TRPA1 to Promote Oxidative-Stress Tolerance. Cancer Cell 33, 985–1003 e1007, doi: 10.1016/j.ccell.2018.05.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schonthal AH Endoplasmic reticulum stress and autophagy as targets for cancer therapy. Cancer Lett 275, 163–169, doi: 10.1016/j.canlet.2008.07.005 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Chen X. & Cubillos-Ruiz JR Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat Rev Cancer 21, 71–88, doi: 10.1038/s41568-020-00312-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eritja N. et al. Autophagy orchestrates adaptive responses to targeted therapy in endometrial cancer. Autophagy 13, 608–624, doi: 10.1080/15548627.2016.1271512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Makhov P. et al. The convergent roles of NF-kappaB and ER stress in sunitinibmediated expression of pro-tumorigenic cytokines and refractory phenotype in renal cell carcinoma. Cell Death Dis 9, 374, doi: 10.1038/s41419-018-0388-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adomavicius T. et al. The structural basis of translational control by eIF2 phosphorylation. Nat Commun 10, 2136, doi: 10.1038/s41467-019-10167-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pavitt GD, Ramaiah KV, Kimball SR & Hinnebusch AG eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev 12, 514–526, doi: 10.1101/gad.12.4.514 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian X. et al. Targeting the Integrated Stress Response in Cancer Therapy. Front Pharmacol 12, 747837, doi: 10.3389/fphar.2021.747837 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spranger S. & Gajewski TF Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer 18, 139–147, doi: 10.1038/nrc.2017.117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang T. et al. BRAF Inhibition Stimulates Melanoma-Associated Macrophages to Drive Tumor Growth. Clin Cancer Res 21, 1652–1664, doi: 10.1158/1078-0432.CCR-14-1554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hipp MM et al. Sorafenib, but not sunitinib, affects function of dendritic cells and induction of primary immune responses. Blood 111, 5610–5620, doi: 10.1182/blood-2007-02-075945 (2008). [DOI] [PubMed] [Google Scholar]
- 92.MEK Inhibition Acts on CD8(+) T Cells to Enhance Antitumor Immunity. Cancer Discov 11, 221, doi: 10.1158/2159-8290.CD-RW2020-176 (2021). [DOI] [Google Scholar]
- 93.Petroni G, Buque A, Zitvogel L, Kroemer G. & Galluzzi L. Immunomodulation by targeted anticancer agents. Cancer Cell 39, 310–345, doi: 10.1016/j.ccell.2020.11.009 (2021). [DOI] [PubMed] [Google Scholar]
- 94.Kersh AE et al. Targeted Therapies: Immunologic Effects and Potential Applications Outside of Cancer. J Clin Pharmacol 58, 7–24, doi: 10.1002/jcph.1028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lampert EJ et al. Combination of PARP Inhibitor Olaparib, and PD-L1 Inhibitor Durvalumab, in Recurrent Ovarian Cancer: a Proof-of-Concept Phase II Study. Clin Cancer Res 26, 4268–4279, doi: 10.1158/1078-0432.CCR-20-0056 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Labrie M. et al. Multiomics analysis of serial PARP inhibitor treated metastatic TNBC inform on rational combination therapies. NPJ Precis Oncol 5, 92, doi: 10.1038/s41698-021-00232-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gaggianesi M. et al. Messing Up the Cancer Stem Cell Chemoresistance Mechanisms Supported by Tumor Microenvironment. Front Oncol 11, 702642, doi: 10.3389/fonc.2021.702642 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 16, 582598, doi: 10.1038/nrc.2016.73 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Sahai E. et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 20, 174–186, doi: 10.1038/s41568-019-0238-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garvey CM et al. Anti-EGFR Therapy Induces EGF Secretion by Cancer-Associated Fibroblasts to Confer Colorectal Cancer Chemoresistance. Cancers (Basel) 12, doi: 10.3390/cancers12061393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Straussman R. et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504, doi: 10.1038/nature11183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilson TR et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505–509, doi: 10.1038/nature11249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zervantonakis IK et al. Fibroblast-tumor cell signaling limits HER2 kinase therapy response via activation of MTOR and antiapoptotic pathways. Proc Natl Acad Sci U S A 117, 16500–16508, doi: 10.1073/pnas.2000648117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lotti F. et al. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J Exp Med 210, 2851–2872, doi: 10.1084/jem.20131195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohuchida K. et al. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor-stromal interactions. Cancer Res 64, 3215–3222, doi: 10.1158/0008-5472.can-03-2464 (2004). [DOI] [PubMed] [Google Scholar]
- 106.Hirata E. et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin beta1/FAK signaling. Cancer Cell 27, 574–588, doi: 10.1016/j.ccell.2015.03.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marusyk A. et al. Spatial Proximity to Fibroblasts Impacts Molecular Features and Therapeutic Sensitivity of Breast Cancer Cells Influencing Clinical Outcomes. Cancer Res 76, 6495–6506, doi: 10.1158/0008-5472.CAN-16-1457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zoeller JJ, Bronson RT, Selfors LM, Mills GB & Brugge JS Niche-localized tumor cells are protected from HER2-targeted therapy via upregulation of an antiapoptotic program in vivo. NPJ Breast Cancer 3, 18, doi: 10.1038/s41523-017-0020-z (2017). This research demonstrates that different microenvironmental niches can influence tumor cells response to HER2 inhibition.
- 109.Benavente S, Sanchez-Garcia A, Naches S, ME LL & Lorente J. Therapy-Induced Modulation of the Tumor Microenvironment: New Opportunities for Cancer Therapies. Front Oncol 10, 582884, doi: 10.3389/fonc.2020.582884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Costa C. et al. Measurement of PIP3 levels reveals an unexpected role for p110beta in early adaptive responses to p110alpha-specific inhibitors in luminal breast cancer. Cancer Cell 27, 97–108, doi: 10.1016/j.ccell.2014.11.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. O’Reilly KE et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 66, 1500–1508, doi: 10.1158/0008-5472.CAN-05-2925 (2006). This research shows the existence of a feedback down-regulation of RTK signaling in presence of a constitutive mTOR activation. This feedback loop is reversed in the presence of an mTOR inhibitor, attenuating its therapeutic effects.
- 112.Schwill M. et al. Systemic analysis of tyrosine kinase signaling reveals a common adaptive response program in a HER2-positive breast cancer. Sci Signal 12, doi: 10.1126/scisignal.aau2875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Brown WS et al. Overcoming Adaptive Resistance to KRAS and MEK Inhibitors by Co-targeting mTORC1/2 Complexes in Pancreatic Cancer. Cell Rep Med 1, 100131, doi: 10.1016/j.xcrm.2020.100131 (2020). This study shows that inhibition of mTORC1/2 results in a compensatory increase in Erk activation, which can be interdicted through MEK and mTORC1/2 dual inhibition, resulting in synergistic activity.
- 114.Shi H. et al. A novel AKT1 mutant amplifies an adaptive melanoma response to BRAF inhibition. Cancer Discov 4, 69–79, doi: 10.1158/2159-8290.CD-13-0279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hanker AB, Kaklamani V. & Arteaga CL Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov 9, 482–491, doi: 10.1158/2159-8290.CD-18-1175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Anderson GR et al. PIK3CA mutations enable targeting of a breast tumor dependency through mTOR-mediated MCL-1 translation. Sci Transl Med 8, 369ra175, doi: 10.1126/scitranslmed.aae0348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hata AN et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 22, 262–269, doi: 10.1038/nm.4040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iavarone C. et al. Combined MEK and BCL-2/XL Inhibition Is Effective in High-Grade Serous Ovarian Cancer Patient-Derived Xenograft Models and BIM Levels Are Predictive of Responsiveness. Mol Cancer Ther 18, 642–655, doi: 10.1158/15357163.MCT-18-0413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Montero J. et al. Destabilization of NOXA mRNA as a common resistance mechanism to targeted therapies. Nat Commun 10, 5157, doi: 10.1038/s41467-019-12477-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zoeller JJ et al. Navitoclax enhances the effectiveness of EGFR-targeted antibodydrug conjugates in PDX models of EGFR-expressing triple-negative breast cancer. Breast Cancer Res 22, 132, doi: 10.1186/s13058-020-01374-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Menon DR et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene 34, 4545, doi: 10.1038/onc.2014.432 (2015). [DOI] [PubMed] [Google Scholar]
- 122.Emran AA et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget 9, 8206–8222, doi: 10.18632/oncotarget.23654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Stuhlmiller TJ et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep 11, 390–404, doi: 10.1016/j.celrep.2015.03.037 (2015). This study demonstrates that inhibition of chromatin readers prevents the kinome reprogramming during the adaptive responses to ERBB2 inhibitor lapatinib.
- 124.Rusan M. et al. Suppression of Adaptive Responses to Targeted Cancer Therapy by Transcriptional Repression. Cancer Discov 8, 59–73, doi: 10.1158/2159-8290.CD-17-0461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Subramanian S, Bates SE, Wright JJ, Espinoza-Delgado I. & Piekarz RL Clinical Toxicities of Histone Deacetylase Inhibitors. Pharmaceuticals (Basel) 3, 2751–2767, doi: 10.3390/ph3092751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Labrie M. et al. Adaptive responses in a PARP inhibitor window of opportunity trial illustrate limited functional interlesional heterogeneity and potential combination therapy options. Oncotarget 10, 3533–3546, doi: 10.18632/oncotarget.26947 (2019). This study demonstrates that adaptive response is detected early in ovarian cancer patients treated with PARP inhibitor and that the response is conserved across multiple lesions from the same patient.
- 127.Lloyd RL et al. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene 39, 4869–4883, doi: 10.1038/s41388-020-1328-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yazinski SA et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 31, 318–332, doi: 10.1101/gad.290957.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y. et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res 61, 8211–8217 (2001). [PubMed] [Google Scholar]
- 130.Sun C, Fang Y, Labrie M, Li X. & Mills GB Systems approach to rational combination therapy: PARP inhibitors. Biochem Soc Trans 48, 1101–1108, doi: 10.1042/BST20191092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ewald B, Sampath D. & Plunkett W. Nucleoside analogs: molecular mechanisms signaling cell death. Oncogene 27, 6522–6537, doi: 10.1038/onc.2008.316 (2008). [DOI] [PubMed] [Google Scholar]
- 132. Fang Y. et al. Sequential Therapy with PARP and WEE1 Inhibitors Minimizes Toxicity while Maintaining Efficacy. Cancer Cell 35, 851–867 e857, doi: 10.1016/j.ccell.2019.05.001 (2019). This research shows that sequential therapy with PARP and WEE1 inhibitors have synergistic activity, while minimizing toxicity. It also illustrates the concept of therapeutic opportunities based on the pre-existing replication stress in cancer cells.
- 133.Aarts M. et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov 2, 524–539, doi: 10.1158/2159-8290.CD-11-0320 (2012). [DOI] [PubMed] [Google Scholar]
- 134.Abildgaard C. & Guldberg P. Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol Med 21, 164–171, doi: 10.1016/j.molmed.2014.12.007 (2015). [DOI] [PubMed] [Google Scholar]
- 135.Kluza J. et al. Inactivation of the HIF-1alpha/PDK3 signaling axis drives melanoma toward mitochondrial oxidative metabolism and potentiates the therapeutic activity of pro-oxidants. Cancer Res 72, 5035–5047, doi: 10.1158/0008-5472.CAN-12-0979 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Sarmiento-Salinas FL et al. Reactive oxygen species: Role in carcinogenesis, cancer cell signaling and tumor progression. Life Sci 284, 119942, doi: 10.1016/j.lfs.2021.119942 (2021). [DOI] [PubMed] [Google Scholar]
- 137.Aggarwal V. et al. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 9, doi: 10.3390/biom9110735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tarrado-Castellarnau M. et al. De novo MYC addiction as an adaptive response of cancer cells to CDK4/6 inhibition. Mol Syst Biol 13, 940, doi: 10.15252/msb.20167321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cuadrado A. et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov 18, 295–317, doi: 10.1038/s41573-018-0008-x (2019). [DOI] [PubMed] [Google Scholar]
- 140.Eluard B, Thieblemont C. & Baud V. NF-kappaB in the New Era of Cancer Therapy. Trends Cancer 6, 677–687, doi: 10.1016/j.trecan.2020.04.003 (2020). [DOI] [PubMed] [Google Scholar]
- 141.Jaganjac M, Milkovic L, Sunjic SB & Zarkovic N. The NRF2, Thioredoxin, and Glutathione System in Tumorigenesis and Anticancer Therapies. Antioxidants (Basel) 9, doi: 10.3390/antiox9111151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang H, Zhang S, Song L, Qu M. & Zou Z. Synergistic lethality between PARPtrapping and alantolactone-induced oxidative DNA damage in homologous recombination-proficient cancer cells. Oncogene 39, 2905–2920, doi: 10.1038/s41388-020-1191-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Buisson R, Boisvert JL, Benes CH & Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell 59, 10111024, doi: 10.1016/j.molcel.2015.07.029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim H. et al. Sulfiredoxin inhibitor induces preferential death of cancer cells through reactive oxygen species-mediated mitochondrial damage. Free Radic Biol Med 91, 264–274, doi: 10.1016/j.freeradbiomed.2015.12.023 (2016). [DOI] [PubMed] [Google Scholar]
- 145.Trachootham D, Alexandre J. & Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 8, 579–591, doi: 10.1038/nrd2803 (2009). [DOI] [PubMed] [Google Scholar]
- 146.Ubhi T. & Brown GW Exploiting DNA Replication Stress for Cancer Treatment. Cancer Res 79, 1730–1739, doi: 10.1158/0008-5472.CAN-18-3631 (2019). [DOI] [PubMed] [Google Scholar]
- 147.Spencer SL & Sorger PK Measuring and modeling apoptosis in single cells. Cell 144, 926–939, doi: 10.1016/j.cell.2011.03.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jiang Y. et al. PARP inhibitors synergize with gemcitabine by potentiating DNA damage in non-small-cell lung cancer. Int J Cancer 144, 1092–1103, doi: 10.1002/ijc.31770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee HJ et al. Combining PARP-1 inhibition and radiation in Ewing sarcoma results in lethal DNA damage. Mol Cancer Ther 12, 2591–2600, doi: 10.1158/1535-7163.MCT-130338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 150.Green DR & Llambi F. Cell Death Signaling. Cold Spring Harb Perspect Biol 7, doi: 10.1101/cshperspect.a006080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang YC et al. Targeting endoplasmic reticulum stress and Akt with OSU-03012 and gefitinib or erlotinib to overcome resistance to epidermal growth factor receptor inhibitors. Cancer Res 68, 2820–2830, doi: 10.1158/0008-5472.CAN-07-1336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mehta AK et al. Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer. Nat Cancer 2, 66–82, doi: 10.1038/s43018-020-00148-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Z. et al. Niraparib activates interferon signaling and synergizes therapeutically with anti-PD-1 antibody. . Scientific Reports, In Press (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pantelidou C. et al. PARP Inhibitor Efficacy Depends on CD8(+) T-cell Recruitment via Intratumoral STING Pathway Activation in BRCA-Deficient Models of Triple-Negative Breast Cancer. Cancer Discov 9, 722–737, doi: 10.1158/2159-8290.CD-18-1218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Farkkila A. et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat Commun 11, 1459, doi: 10.1038/s41467-02015315-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang B. et al. MEK inhibition remodels the immune landscape of mutant KRAS tumors to overcome resistance to PARP and immune checkpoint inhibitors. Cancer Res, doi: 10.1158/0008-5472.CAN-20-2370 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jain RK Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science 307, 58–62, doi: 10.1126/science.1104819 (2005). [DOI] [PubMed] [Google Scholar]
- 158. Chauhan VP et al. Reprogramming the microenvironment with tumor-selective angiotensin blockers enhances cancer immunotherapy. Proc Natl Acad Sci U S A 116, 10674–10680, doi: 10.1073/pnas.1819889116 (2019). This study demonstrates that angiotensin receptor blockers can reprogram cancer-associated fibroblasts to promote T-cell activity and enhance immunotherapy.
- 159.Murphy JE et al. Total Neoadjuvant Therapy With FOLFIRINOX in Combination With Losartan Followed by Chemoradiotherapy for Locally Advanced Pancreatic Cancer: A Phase 2 Clinical Trial. JAMA Oncol 5, 1020–1027, doi: 10.1001/jamaoncol.2019.0892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Alishekevitz D. et al. Macrophage-Induced Lymphangiogenesis and Metastasis following Paclitaxel Chemotherapy Is Regulated by VEGFR3. Cell Rep 17, 1344–1356, doi: 10.1016/j.celrep.2016.09.083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Apte RS, Chen DS & Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 176, 1248–1264, doi: 10.1016/j.cell.2019.01.021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tang C. et al. Synergy between VEGF/VEGFR inhibitors and chemotherapy agents in the phase I clinic. Clin Cancer Res 20, 5956–5963, doi: 10.1158/1078-0432.CCR-141582 (2014). [DOI] [PubMed] [Google Scholar]
- 163.Gilad Y, Gellerman G, Lonard DM & O’Malley BW Drug Combination in Cancer Treatment-From Cocktails to Conjugated Combinations. Cancers (Basel) 13, doi: 10.3390/cancers13040669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kircher MF, Hricak H. & Larson SM Molecular imaging for personalized cancer care. Mol Oncol 6, 182–195, doi: 10.1016/j.molonc.2012.02.005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mankoff DA, Farwell MD, Clark AS & Pryma DA Making Molecular Imaging a Clinical Tool for Precision Oncology: A Review. JAMA Oncol 3, 695–701, doi: 10.1001/jamaoncol.2016.5084 (2017). [DOI] [PubMed] [Google Scholar]
- 166.de Vries EGE et al. Integrating molecular nuclear imaging in clinical research to improve anticancer therapy. Nat Rev Clin Oncol 16, 241–255, doi: 10.1038/s41571-0180123-y (2019). [DOI] [PubMed] [Google Scholar]
- 167.Kilgour E, Rothwell DG, Brady G. & Dive C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 37, 485–495, doi: 10.1016/j.ccell.2020.03.012 (2020). [DOI] [PubMed] [Google Scholar]
- 168.Rodriguez J. et al. When Tissue is an Issue the Liquid Biopsy is Nonissue: A Review. Oncol Ther 9, 89–110, doi: 10.1007/s40487-021-00144-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu J. et al. Circulating Tumor Cells (CTCs): A Unique Model of Cancer Metastases and Non-invasive Biomarkers of Therapeutic Response. Front Genet 12, 734595, doi: 10.3389/fgene.2021.734595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sanjabi S. & Lear S. New cytometry tools for immune monitoring during cancer immunotherapy. Cytometry B Clin Cytom 100, 10–18, doi: 10.1002/cyto.b.21984 (2021). [DOI] [PubMed] [Google Scholar]
- 171.Hernandez C. et al. Systemic Blood Immune Cell Populations as Biomarkers for the Outcome of Immune Checkpoint Inhibitor Therapies. Int J Mol Sci 21, doi: 10.3390/ijms21072411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Reduzzi C. et al. The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Semin Cancer Biol 60, 344–350, doi: 10.1016/j.semcancer.2019.10.008 (2020). [DOI] [PubMed] [Google Scholar]
- 173.Martinez-Dominguez MV et al. Current Technologies for RNA-Directed Liquid Diagnostics. Cancers (Basel) 13, doi: 10.3390/cancers13205060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Martellucci S. et al. Extracellular Vesicles: New Endogenous Shuttles for miRNAs in Cancer Diagnosis and Therapy? Int J Mol Sci 21, doi: 10.3390/ijms21186486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Valencia K. & Montuenga LM Exosomes in Liquid Biopsy: The Nanometric World in the Pursuit of Precision Oncology. Cancers (Basel) 13, doi: 10.3390/cancers13092147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Di Martino MT et al. miRNAs and lncRNAs as Novel Therapeutic Targets to Improve Cancer Immunotherapy. Cancers (Basel) 13, doi: 10.3390/cancers13071587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gheibi-Hayat SM & Jamialahmadi K. Antisense Oligonucleotide (AS-ODN) Technology: Principle, Mechanism and Challenges. Biotechnol Appl Biochem, doi: 10.1002/bab.2028 (2020). [DOI] [PubMed] [Google Scholar]
- 178.Romano G, Acunzo M. & Nana-Sinkam P. microRNAs as Novel Therapeutics in Cancer. Cancers (Basel) 13, doi: 10.3390/cancers13071526 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Belete TM The Current Status of Gene Therapy for the Treatment of Cancer. Biologics 15, 67–77, doi: 10.2147/BTT.S302095 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Canon J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223, doi: 10.1038/s41586-019-1694-1 (2019). [DOI] [PubMed] [Google Scholar]
- 181.Hallin J. et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 10, 54–71, doi: 10.1158/2159-8290.CD-19-1167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Mitri ZI et al. Implementing a comprehensive translational oncology platform: from molecular testing to actionability. J Transl Med 16, 358, doi: 10.1186/s12967-018-1733-y (2018). This study shows the feasibility of real-time cancer tissue acquisition and analysis within a translational oncology platform that aims at providing biomarker driven precision therapy to cancer patients.
- 183.Janiszewska M. et al. The impact of tumor epithelial and microenvironmental heterogeneity on treatment responses in HER2+ breast cancer. JCI Insight 6, doi: 10.1172/jci.insight.147617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Roper N. et al. Clonal Evolution and Heterogeneity of Osimertinib Acquired Resistance Mechanisms in EGFR Mutant Lung Cancer. Cell Rep Med 1, doi: 10.1016/j.xcrm.2020.100007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Marin-Bejar O. et al. Evolutionary predictability of genetic versus nongenetic resistance to anticancer drugs in melanoma. Cancer Cell 39, 1135–1149 e1138, doi: 10.1016/j.ccell.2021.05.015 (2021). [DOI] [PubMed] [Google Scholar]
- 186.Guruprasad P, Lee YG, Kim KH & Ruella M. The current landscape of single-cell transcriptomics for cancer immunotherapy. J Exp Med 218, doi: 10.1084/jem.20201574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lim B, Lin Y. & Navin N. Advancing Cancer Research and Medicine with Single-Cell Genomics. Cancer Cell 37, 456–470, doi: 10.1016/j.ccell.2020.03.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Paul I, White C, Turcinovic I. & Emili A. Imaging the future: the emerging era of single-cell spatial proteomics. FEBS J, doi: 10.1111/febs.15685 (2020). [DOI] [PubMed] [Google Scholar]
- 189.Hegde PS & Chen DS Top 10 Challenges in Cancer Immunotherapy. Immunity 52, 17–35, doi: 10.1016/j.immuni.2019.12.011 (2020). [DOI] [PubMed] [Google Scholar]
- 190.Wargo JA, Reuben A, Cooper ZA, Oh KS & Sullivan RJ Immune Effects of Chemotherapy, Radiation, and Targeted Therapy and Opportunities for Combination With Immunotherapy. Semin Oncol 42, 601–616, doi: 10.1053/j.seminoncol.2015.05.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Miller MA & Weissleder R. Imaging of anticancer drug action in single cells. Nat Rev Cancer 17, 399–414, doi: 10.1038/nrc.2017.41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Salvador-Barbero B. et al. CDK4/6 Inhibitors Impair Recovery from Cytotoxic Chemotherapy in Pancreatic Adenocarcinoma. Cancer Cell 38, 584, doi: 10.1016/j.ccell.2020.09.012 (2020). [DOI] [PubMed] [Google Scholar]
- 193.Capivasertib Active against AKT1-Mutated Cancers. Cancer Discov 9, OF7, doi: 10.1158/2159-8290.CD-NB2018-153 (2019). [DOI] [PubMed] [Google Scholar]
- 194.Gnann C, Cesnik AJ & Lundberg E. Illuminating Non-genetic Cellular Heterogeneity with Imaging-Based Spatial Proteomics. Trends Cancer 7, 278–282, doi: 10.1016/j.trecan.2020.12.006 (2021). [DOI] [PubMed] [Google Scholar]
- 195.Drost J. & Clevers H. Organoids in cancer research. Nat Rev Cancer 18, 407–418, doi: 10.1038/s41568-018-0007-6 (2018). [DOI] [PubMed] [Google Scholar]
- 196.Gengenbacher N, Singhal M. & Augustin HG Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat Rev Cancer 17, 751–765, doi: 10.1038/nrc.2017.92 (2017). [DOI] [PubMed] [Google Scholar]
- 197.Sontheimer-Phelps A, Hassell BA & Ingber DE Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer 19, 65–81, doi: 10.1038/s41568-018-0104-6 (2019). [DOI] [PubMed] [Google Scholar]
- 198.Morgan D, Jost TA, De Santiago C. & Brock A. Applications of high-resolution clone tracking technologies in cancer. Current Opinion in Biomedical Engineering 19, doi: 10.1016/j.cobme.2021.100317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Saavedra-Garcia P. et al. Systems level profiling of chemotherapy-induced stress resolution in cancer cells reveals druggable trade-offs. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2018229118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhao W. et al. Large-Scale Characterization of Drug Responses of Clinically Relevant Proteins in Cancer Cell Lines. Cancer Cell 38, 829–843 e824, doi: 10.1016/j.ccell.2020.10.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Avolio R, Matassa DS, Criscuolo D, Landriscina M. & Esposito F. Modulation of Mitochondrial Metabolic Reprogramming and Oxidative Stress to Overcome Chemoresistance in Cancer. Biomolecules 10, doi: 10.3390/biom10010135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Loponte S, Lovisa S, Deem AK, Carugo A. & Viale A. The Many Facets of Tumor Heterogeneity: Is Metabolism Lagging Behind? Cancers (Basel) 11, doi: 10.3390/cancers11101574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Singh B, Patwardhan RS, Jayakumar S, Sharma D. & Sandur SK Oxidative stress associated metabolic adaptations regulate radioresistance in human lung cancer cells. J Photochem Photobiol B 213, 112080, doi: 10.1016/j.jphotobiol.2020.112080 (2020). [DOI] [PubMed] [Google Scholar]
- 204.Tu W, Wang H, Li S, Liu Q. & Sha H. The Anti-Inflammatory and Anti-Oxidant Mechanisms of the Keap1/Nrf2/ARE Signaling Pathway in Chronic Diseases. Aging Dis 10, 637–651, doi: 10.14336/AD.2018.0513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ju HQ et al. Mechanisms of Overcoming Intrinsic Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma through the Redox Modulation. Mol Cancer Ther 14, 788–798, doi: 10.1158/1535-7163.MCT-14-0420 (2015). [DOI] [PubMed] [Google Scholar]
- 206.Xu J, Patel NH & Gewirtz DA Triangular Relationship between p53, Autophagy, and Chemotherapy Resistance. Int J Mol Sci 21, doi: 10.3390/ijms21238991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Deng D. & Shah K. TRAIL of Hope Meeting Resistance in Cancer. Trends Cancer 6, 989–1001, doi: 10.1016/j.trecan.2020.06.006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Stohr D, Jeltsch A. & Rehm M. TRAIL receptor signaling: From the basics of canonical signal transduction toward its entanglement with ER stress and the unfolded protein response. Int Rev Cell Mol Biol 351, 57–99, doi: 10.1016/bs.ircmb.2020.02.002 (2020). [DOI] [PubMed] [Google Scholar]
- 209.Hatok J. & Racay P. Bcl-2 family proteins: master regulators of cell survival. Biomol Concepts 7, 259–270, doi: 10.1515/bmc-2016-0015 (2016). [DOI] [PubMed] [Google Scholar]
- 210.Lemmer IL, Willemsen N, Hilal N. & Bartelt A. A guide to understanding endoplasmic reticulum stress in metabolic disorders. Mol Metab 47, 101169, doi: 10.1016/j.molmet.2021.101169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Akman M. et al. Hypoxia, endoplasmic reticulum stress and chemoresistance: dangerous liaisons. J Exp Clin Cancer Res 40, 28, doi: 10.1186/s13046-020-01824-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jing X. et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer 18, 157, doi: 10.1186/s12943-019-1089-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Samanta D. & Semenza GL Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochim Biophys Acta Rev Cancer 1870, 15–22, doi: 10.1016/j.bbcan.2018.07.002 (2018). [DOI] [PubMed] [Google Scholar]
- 214.Baillie KE & Stirling PC Beyond Kinases: Targeting Replication Stress Proteins in Cancer Therapy. Trends Cancer 7, 430–446, doi: 10.1016/j.trecan.2020.10.010 (2021). [DOI] [PubMed] [Google Scholar]
- 215.Gmeiner WH Entrapment of DNA topoisomerase-DNA complexes by nucleotide/nucleoside analogs. Cancer Drug Resist 2, 994–1001, doi: 10.20517/cdr.2019.95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pilie PG, Tang C, Mills GB & Yap TA State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol 16, 81–104, doi: 10.1038/s41571018-0114-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rebe C. & Ghiringhelli F. STAT3, a Master Regulator of Anti-Tumor Immune Response. Cancers (Basel) 11, doi: 10.3390/cancers11091280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vanpouille-Box C. et al. TGFbeta Is a Master Regulator of Radiation Therapy-Induced Antitumor Immunity. Cancer Res 75, 2232–2242, doi: 10.1158/0008-5472.CAN-14-3511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Alcala AM & Flaherty KT BRAF inhibitors for the treatment of metastatic melanoma: clinical trials and mechanisms of resistance. Clin Cancer Res 18, 33–39, doi: 10.1158/1078-0432.CCR-11-0997 (2012). [DOI] [PubMed] [Google Scholar]
- 220.Carracedo A. et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest 118, 3065–3074, doi: 10.1172/JCI34739 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zoeller JJ et al. Clinical evaluation of BCL-2/XL levels pre- and post- HER2-targeted therapy. PLoS One 16, e0251163, doi: 10.1371/journal.pone.0251163 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bosch A. et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 7, 283ra251, doi: 10.1126/scitranslmed.aaa4442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Leone A. et al. Vorinostat synergizes with EGFR inhibitors in NSCLC cells by increasing ROS via up-regulation of the major mitochondrial porin VDAC1 and modulation of the cMyc-NRF2-KEAP1 pathway. Free Radic Biol Med 89, 287–299, doi: 10.1016/j.freeradbiomed.2015.07.155 (2015). [DOI] [PubMed] [Google Scholar]
- 224.Boucher Y. et al. Bevacizumab improves tumor infiltration of mature dendritic cells and effector T-cells in triple-negative breast cancer patients. NPJ Precis Oncol 5, 62, doi: 10.1038/s41698-021-00197-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zervantonakis IK et al. Systems analysis of apoptotic priming in ovarian cancer identifies vulnerabilities and predictors of drug response. Nat Commun 8, 365, doi: 10.1038/s41467-017-00263-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Leonard B. et al. BET Inhibition Overcomes Receptor Tyrosine Kinase-Mediated Cetuximab Resistance in HNSCC. Cancer Res 78, 4331–4343, doi: 10.1158/00085472.CAN-18-0459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zawistowski JS et al. Enhancer Remodeling during Adaptive Bypass to MEK Inhibition Is Attenuated by Pharmacologic Targeting of the P-TEFb Complex. Cancer Discov 7, 302–321, doi: 10.1158/2159-8290.CD-16-0653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nath A. & Bild AH Leveraging Single-Cell Approaches in Cancer Precision Medicine. Trends Cancer 7, 359–372, doi: 10.1016/j.trecan.2021.01.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Califano A. & Alvarez MJ The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer 17, 116–130, doi: 10.1038/nrc.2016.124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Schwarz RF et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med 12, e1001789, doi: 10.1371/journal.pmed.1001789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zhang AW et al. Interfaces of Malignant and Immunologic Clonal Dynamics in Ovarian Cancer. Cell 173, 1755–1769 e1722, doi: 10.1016/j.cell.2018.03.073 (2018). [DOI] [PubMed] [Google Scholar]
- 232.Howard SC, Jones DP & Pui CH The tumor lysis syndrome. N Engl J Med 364, 1844–1854, doi: 10.1056/NEJMra0904569 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Infante JR et al. A phase I dose-escalation study of Selumetinib in combination with Erlotinib or Temsirolimus in patients with advanced solid tumors. Invest New Drugs 35, 576–588, doi: 10.1007/s10637-017-0459-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Senapati S, Mahanta AK, Kumar S. & Maiti P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct Target Ther 3, 7, doi: 10.1038/s41392-017-0004-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]