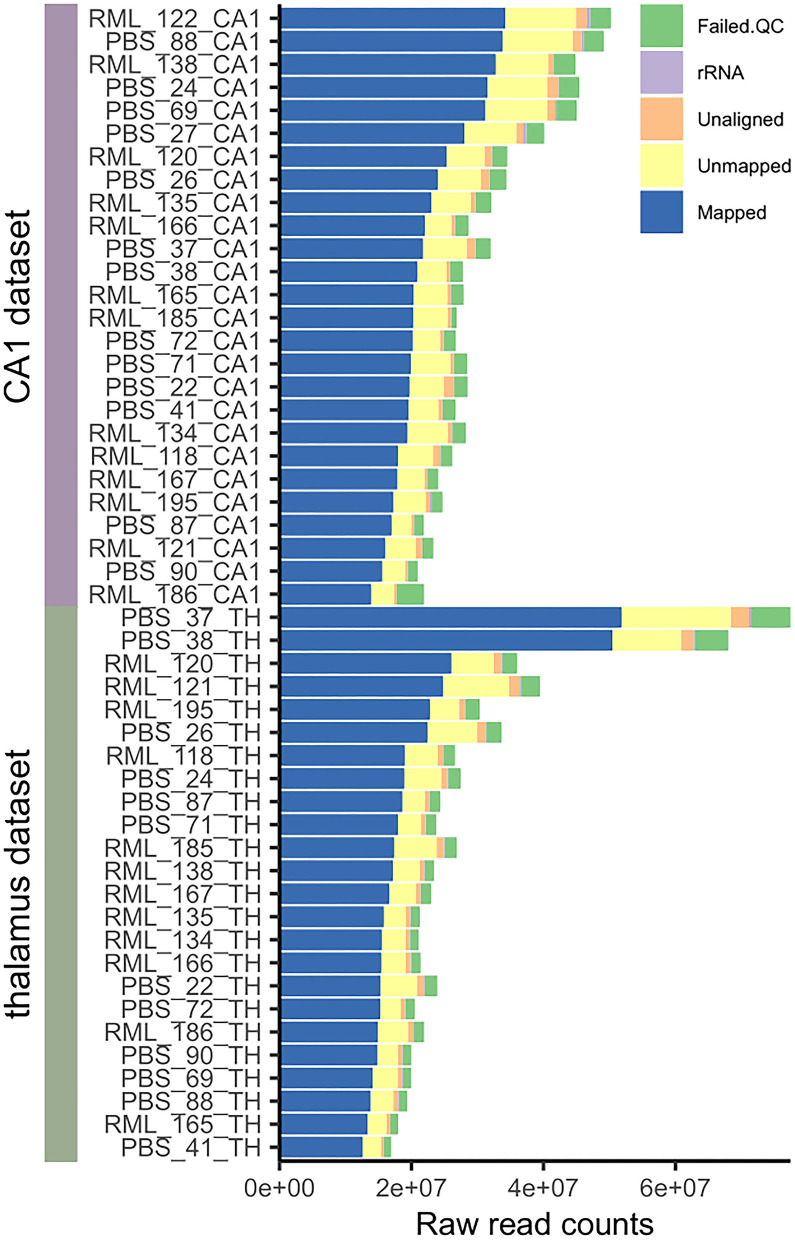
Figure 1.
Pre-processing mapping statistics of all microdissected CA1 and thalamus samples used in the analysis. The number of raw sequencing reads that failed quality control, were removed on the basis of mapping to rRNA, were not aligned to the genome, failed to map to a valid transcript, and were successfully mapped to a transcript are indicated. We aimed for 30–40 million raw sequencing read pairs per library and successfully mapped 15–30 million read pairs per library for the analysis.