Abstract
Myeloid-derived suppressor cells (MDSCs) are a compendium of immature myeloid cells that exhibit potent T-cell suppressive capacity and expand during pathological conditions such as cancer and chronic infections. Although well-characterized in cancer, the physiology of MDSCs in the infection setting remains enigmatic. Here, we integrated single-cell RNA sequencing (scRNA-seq) and functional metabolic profiling to gain deeper insights into the factors governing the generation and maintenance of MDSCs in chronic Staphylococcus aureus infection. We found that MDSCs originate not only in the bone marrow but also at extramedullary sites in S. aureus-infected mice. scRNA-seq showed that infection-driven MDSCs encompass a spectrum of myeloid precursors in different stages of differentiation, ranging from promyelocytes to mature neutrophils. Furthermore, the scRNA-seq analysis has also uncovered valuable phenotypic markers to distinguish mature myeloid cells from immature MDSCs. Metabolic profiling indicates that MDSCs exhibit high glycolytic activity and high glucose consumption rates, which are required for undergoing terminal maturation. However, rapid glucose consumption by MDSCs added to infection-induced perturbations in the glucose supplies in infected mice hinders the terminal maturation of MDSCs and promotes their accumulation in an immature stage. In a proof-of-concept in vivo experiment, we demonstrate the beneficial effect of increasing glucose availability in promoting MDSC terminal differentiation in infected mice. Our results provide valuable information of how metabolic alterations induced by infection influence reprogramming and differentiation of MDSCs.
Keywords: Myeloid-derived suppressor cells, Staphylococcus aureus, Immunometabolism, Single-cell RNA sequencing
Introduction
MDSCs are an aberrant population of immature myeloid cells that fail to undergo terminal differentiation and accumulate during pathological conditions such as cancer, chronic infection, and autoimmunity [1, 2]. In contrast to normal, mature myeloid cells, which play a pivotal role in host defense against pathogens and in the initiation of T-cell immunity, MDSCs exert immune regulatory functions and are potent suppressors of T-cell responses [3]. In humans and mice, MDSCs have been typically divided into 2 different subsets, monocytic and granulocytic, based on cell surface markers. In humans, granulocytic MDSCs are identified by the expression of CD15+CD11b+CD33+HLA-DR− and monocytic MDSCs by the expression of CD14+CD11b+CD33+HLA-DR− [4]. In mice, monocytic MDSCs express CD11b+Ly6C+Ly6Glow, while granulocytic MDSCs express CD11b+Ly6ClowL6G+ [4]. However, it has become clear that this classification is rather simplistic and does not recapitulate the high degree of phenotypic and functional heterogeneity of MDSCs [5, 6].
Although MDSCs have been extensively studied and characterized in the cancer setting, where they seem to play an important role in supporting tumor progression [7], mounting evidence indicates that MDSCs play also an important regulatory role in the immune response to pathogens [8]. MDSCs have been reported to play an important role in chronic infections caused by S. aureus [9, 10, 11, 12, 13, 14], which is a major human pathogen that causes a wide variety of infections ranging from mild, self-limited infections to chronic and difficult-to-treat diseases including osteomyelitis, prosthetic joint infections, and biofilm-related infections [15]. We have previously reported the expansion of MDSCs in murine models of S. aureus chronic abscesses and bone infection where they induce progressive T-cell dysfunction and promote pathogen persistence [12]. In the same line, Heim and colleagues [9, 11, 14] demonstrated that MDSCs infiltrate the site of infection in a mouse model of S. aureus orthopedic implant infection, where they promote an anti-inflammatory environment that favore biofilm persistence. Accumulation of granulocytic MDSCs at the site of prosthetic joint infections has also been observed in humans [13]. The authors proposed that the accumulation of MDSCs could account for the chronicity of these infections [13]. All together, these observations indicate that MDSCs are an important element of the host response to S. aureus chronic infections, and therefore, targeting MDSCs may represent a promising therapeutic intervention to overcome immunosuppression and facilitate pathogen clearance by the immune system.
In cancer, several preclinical and clinical studies have shown the benefit of including MDSC-targeting approaches such as depletion of MDSCs or blockade of MDSCs migration in combination therapies to reduce tumor progression [16]. In the infection setting, however, considering that MDSCs also encompass populations of mature myeloid cells that are critical for the control of many pathogens, these strategies may have a profound negative effect in the course of infection. The development of such strategies will require a better understanding of how MDSCs are generated during chronic infection, which factors are involved in the process, and the mechanisms that prevent their maturation. In the current study, we used single-cell RNA sequencing (scRNA-seq) and metabolic profiling to investigate the origin, heterogeneity, molecular mechanisms, and pathways underlying the development and maintenance of MDSCs in a murine model of S. aureus chronic infection.
Materials and Methods
Bacterial Strains
S. aureus strains 6850 and SH1000 were grown to the mid-log phase in brain heart infusion medium (BHI, Roth) at 37°C with shaking (120 rpm), collected by centrifugation, washed with sterile PBS, and diluted to the required concentration. The number of viable bacteria was determined by tenfold serial dilution and colony count by plating on blood agar.
Mice and Infection Model
Pathogen-free 9- to 10-week-old C57BL/6 female mice were purchased from Envigo (The Netherlands) and maintained according to institutional guidelines in individually ventilated cages with food and water provided ad libitum. Mice were intravenously inoculated either with 106 CFU of S. aureus strain 6850 or with 4 × 107 CFU of S. aureus strain SH1000 in 100 μL of PBS via a lateral tail vein, and sacrificed by CO2 asphyxiation at indicated times. Bacteria were counted in the tibia and spleen by preparing homogenates in PBS and plating tenfold serial dilutions on blood agar. In some experiments, infected mice were fed with water supplemented with 10% glucose during 10 days after bacterial inoculation. This time period was selected to minimize potential secondary metabolic alterations such as increased glucose intolerance and insulin resistance associated with long-term consumption of glucose-sweetened water. Blood glucose was measured using a Contour XT glucometer (Bayer).
Cell suspensions were prepared from the spleen of infected mice by gently teasing the spleen tissue through a 100-µm pore size nylon cell strainer and PBS+10% FCS. Splenocytes were spun down and erythrocytes were lysed after incubation for 5 min at RT in ammonium-chloride-potassium lysing buffer and then washed 3 times in PBS+10% FCS.
The bone marrow was flushed out of both tibia and femur from one hind limb nonaffected by the infection using a 21-gauge needle attached to a 5-mL syringe filled with PBS, followed by centrifugation and erythrocyte removal with ammonium-chloride-potassium.
Flow Cytometry Analysis
Cell suspensions were incubated with anti-mouse CD16/32 (eBioscience) for 5 min at RT to block Fc receptors and stained for 20 min at 4°C with antibodies against surface antigens. Cells were washed with PBS+10% FCS followed by fixation for 15 min with fixation buffer (BioLegend) and analyzed on a LSRII cytometer (Becton Dickinson).
For intracellular staining, cells were stained first against surface antigens as described above, fixed for 15 min at RT with fixation buffer, washed twice with permeabilization buffer (BioLegend), and stained for intracellular markers. After washing with permeabilization buffer, cells were analyzed on a LSRII cytometer. Data were analyzed using FlowJo v9.3 software.
The following antibodies have been used in this study: rat anti-mouse CD11b-PE/Cy7, rat anti-mouse CD45R/B220-PE, rat anti-mouse Ly6C-APC, rat anti-mouse Ly6G-PE, rat anti-mouse Ly6G-APC, anti-mouse CD117 (c-Kit)-FITC, rat anti-mouse Ly6A/E (Sca-1)-APC/Cy7, rat anti-mouse Lineage Cocktail-PB, and rat anti-mouse CD127 (IL-7Rα)-PE/Cy5, all from BioLegend; rat anti-mouse CD4-FITC, rat anti-mouse CD8-PE, and CD16/CD32 unconjugated from eBioscience; Armenian hamster anti-mouse CD3e purified and Syrian hamster anti-mouse CD28 purified from BD Pharmingen; and rat anti-mouse CCL6-Alexa Fluor 647 from R&D Systems.
Cell viability was determined by flow cytometry using propidium iodide solution following the manufacturer's recommendations (BioLegend).
Carboxyfluorescein Succinimidyl Ester Staining and Proliferation Assay
CD4+ T cells were isolated from the spleen of uninfected mice using the mouse CD4+ T Cell Isolation kit (Miltenyi Biotec), and Ly6C+Ly6G+ MDSCs were isolated from the spleen of S. aureus-infected mice at day 21 of infection using the mouse Myeloid-Derived Suppressor Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer's instructions. Isolated CD4+ T cells were then labeled with carboxyfluorescein succinimidyl ester (BioLegend) following the manufacturer's recommendations and cultured at 5 × 105 cells per well in complete RPMI-1640 medium (Gibco) supplemented with antibiotic-antimycotic (1:1,000) (VWR International), 4 mML-glutamine (Sigma-Aldrich), and 10% FCS and 2 μg/mL of Armenian hamster anti-mouse CD3ε plus 2 μg/mL of Syrian hamster anti-mouse CD28 antibodies (BD Pharmingen) at 37°C and 5% CO2 for 72 h in the presence or absence of 5 × 105 per well of MDSCs isolated from the spleen of S. aureus-infected mice at a 1:1 ratio. Unstimulated CD4+ T cells incubated in medium without anti-CD3ε and anti-CD28 antibodies were used as control. Proliferation was determined by flow cytometry analysis and dilution of CSFE as indication of cell division.
Cytokine Determination
IL-2 and IFN-γ levels were determined in the culture supernatant of CD4+ T cells unstimulated or stimulated for 72 h with anti-CD3/anti-CD28 in the presence or absence of MDSCs isolated from the spleen of S. aureus-infected mice at day 21 of infection using mouse IL-2 and mouse IFN-γ ELISA sets according to the manufacturer's recommendations (BD Biosciences).
Histology
Spleens were removed from uninfected or S. aureus-infected mice at day 21 of infection, fixed in 10% formalin, and embedded in paraffin. Tissue section samples (2 μm thick) were stained with hematoxylin/eosin (Roth) and examined under a light microscope.
Single-Cell RNA Sequencing
Spleens isolated from 5 uninfected and 5 S. aureus-infected mice (day 21 of infection) were transformed into a single-cell suspension and pooled. The CD11b+ populations in the infected and uninfected samples were sorted using a FACSAria(TM) SORP and approximately 4,000 cells loaded onto the 10x Genomics Chromium Controller following the single-cell 3′ v3 protocol (10x Genomics). Libraries were prepared from single-cell suspensions according to the 10x Genomics 3′ v3 protocol and sequenced using an Illumina NovaSeq 6000 sequencer (Illumina) with a sequencing depth of 200 million reads per sample.
In vivo 5-Ethynyl-2′-Deoxyuridine-Based Cell Proliferation Assay
5-Ethynyl-2′-deoxyuridine (EdU) (Thermo Fisher Scientific) was administered intraperitoneally (0.5 mg/mice) to uninfected or S. aureus-infected mice (day 21 of infection) 24 h before sacrifice. Spleens were removed, converted into a single-cell suspension, and stained for surface markers. EdU staining was performed with the Click-iT EdU AlexaFluor647 Flow Cytometry Assay Kit following the manufacturer's instructions (Thermo Fisher Scientific). Proliferating cells were determined by flow cytometry analysis.
In vitro Culture of MDSCs
Spleen cells isolated from S. aureus-infected mice (day 21 of infection) were cultured in vitro at a density of 5 × 106 cells/mL in complete RPMI-1640 medium at 37°C, 5% CO2. Cells were harvested at the indicated times of in vitro culture, stained with antibodies against the surface marker Ly6G and with antibodies against the intracellular marker CCL6, and analyzed by flow cytometry.
In some experiments, spleen cells were cultured in complete RPMI-1640 medium containing different concentrations of glucose (0, 0.5, 1, and 2 mg/mL). To inhibit glycolysis, spleen cells were incubated in complete RPMI-1640 medium containing glucose (2 mg/mL) in the presence of 10 mM of the glycolysis inhibitor 2-deoxy-D-glucose (2-DG).
Cytospin
Cytospin were prepared using aliquots of in vitro-cultured MDSCs. The material was centrifuged at 500 rpm for 5 min in a Shandon cytocentrifuge (Cytospin 2, Shandon, UK). Slides were stained using May-Grünwald-Giemsa (Polysciences) and photographed with a light microscope.
Phagocytosis and Killing Assay
Ly6C+Ly6G+ cells were isolated from S. aureus-infected mice (day 21 of infection) and cultured for 96 h in complete RPMI medium. Cells were collected, washed, seeded in multi-well plates at 5 × 105 cells per well, and incubated with S. aureus at an MOI of 10:1 in the presence of 10% mouse serum. The plates were centrifuged at 700 g for 5 min and incubated at 37°C for 1 h to allow phagocytosis. Noningested extracellular bacteria were then killed by addition of 100 μg/mL gentamicin (Gibco) and 5 μg/mL lysostaphin (Sigma-Aldrich), and cells were washed and further incubated for 3 h at 37°C. Cells were then harvested, pelleted by centrifugation, and lysed with 0.1% Triton X-100 (Sigma), and CFU were enumerated by plating on blood agar.
Glucose Uptake Assay
Ly6C+Ly6G+ cells were isolated from S. aureus-infected mice (day 21 of infection) using the Myeloid-Derived Suppressor Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer's instructions. Ly6C+Ly6G+ cells prior to (ex vivo) or after in vitro culture for 96 h (in vitro) were seeded in 48-well plates at 106 cells per well in glucose-free RPMI-1640 medium supplemented with 300 μM 2-(N-[7-nitrobenz-2-oxa-1,3-diazol-4-yl] amino)-2-deoxyglucose (Thermo Fisher Scientific) and incubated for 30 min at 37°C, 5% CO2. Cells were washed and analyzed by flow cytometry.
Seahorse Extracellular Flux Analysis
Ly6C+Ly6G+ cells were isolated from S. aureus-infected mice (day 21 of infection) using the Myeloid-Derived Suppressor Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer's instructions and used prior to (ex vivo) or after in vitro culture for 96 h (in vitro). Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of Ly6C+Ly6G+ cells were assessed using an Agilent Seahorse XF96 Analyzer (Agilent Technologies). One day prior to the assay, the Seahorse XF Utility Plate (Agilent Technologies) was hydrated by adding 200 μL of sterile Milli-Q H2O to each well and incubated overnight in a non-CO2 37°C incubator together with the XF sensor cartridge (Agilent Technologies). Before seeding the cells, the wells of a Seahorse 96-well XF cell culture plate (Agilent Technologies) were incubated with poly-L-lysin (Sigma-Aldrich) for 1 h at 37°C, extensively washed with Milli-Q H2O after removing the poly-L-lysin, and left to dry for 30 min at 37°C. Cells were added to poly-L-lysin-coated plates at a concentration of 5 × 105 cells per well in 180 μL Seahorse RPMI medium supplemented with 10 mM glucose and 2 mM glutamine (pH 7.4) and centrifuged at 1,000 g for 5 min. The wells filled up with only assay medium were used as background control. Water was removed from the wells in the utility plate and 200 μL prewarmed (37°C) Seahorse XF calibrant solution (Agilent Technologies) was added to each well. The cell culture plate and utility plate with the sensor cartridge were equilibrated after incubation in a non-CO2 37°C incubator for 1 h. The different inhibitors of the Seahorse XF Cell Glycolytic Rate Assay Kit (Agilent Technologies) were added to the corresponding ports of the sensor cartridge prior to starting the assay. Thus, 20 μL of 5 μM rotenone and 5 μM antimycin A solution was added to port A and 22 μL of 500 μM 2-DG solution to port B. The utility plate and the sensor cartridge were then placed into the XF96 analyzer and calibrated. After calibration, the utility plate was replaced by the cell culture plate, and cell respiration parameters were determined by stepwise injection of the different inhibitors. During each measurement cycle, the OCR and ECAR were determined 3 times including 3 min of mixing and 3 min of measurement.
Stable Isotope Labeling, Metabolite Extraction, GC-MS Measurement, and Data Processing
Ly6C+Ly6G+ cells isolated from the spleen of S. aureus-infected mice (day 21 of infection) prior to (ex vivo) and after in vitro culture for 96 h (in vitro) were seeded in 6-well plates at 8 × 106 cells per well in RPMI medium containing either 11 mM [U-13C6]-glucose (Cambridge Isotope Laboratories), 2 mM [U-13C5]-glutamine (Cambridge Isotope Laboratories), or 100 μM [U-13C16]-palmitate (Cambridge Isotope Laboratories), 10% FCS, 1% penicillin (10,000 IU/mL), and streptomycin (20 mg/mL). 12C metabolites were added to each tracer to ensure equivalent nutrient state. Before the treatment, [U-13C16]-palmitate was noncovalently conjugated to fatty-acid-free BSA (Sigma-Aldrich) as previously described [17]. The cells were incubated at 37°C, 5% CO2 for 4 h. Cells were then harvested and intracellular metabolites were extracted as previously described [19]. Briefly, cell suspensions were collected and centrifuged at 250 g for 5 min. The cell pellet was washed with 1 mL 0.9% NaCl, followed by centrifugation at 250 g for 5 min. The cells were immediately put on ice to quench the metabolism, and 250 μL ice-cold HPLC-grade methanol (Sigma-Aldrich), 250 μL Milli-Q H2O with 1 μg/mL D6 glutaric acid (CDN isotopes) as internal standard, and 250 μL HPLC-grade chloroform (Sigma-Aldrich) were added. The cells were agitated at 4°C for 20 min at 1,400 rpm, followed by centrifugation at 17,000 g at 4°C for 5 min. After phase separation, 300 μL of the polar phase was transferred to a glass vial with a micro-insert and dried at 4°C under vacuum. Derivatization for gas chromatography was performed using a Gerstel MPS. Dried polar metabolites were dissolved in 15 μL of 20 mg/mL methoxyamine hydrochloride (Sigma-Aldrich) in pyridine (Roth) at 40°C while shaking for 90 min. An equal volume of N-tert-butyldimethylsilyl-N-methytrifluoroacetamide (Restek) was then injected, and the cells were further incubated for 60 min at 55°C under shaking. GC-MS measurement was performed on an Agilent 7890B GC coupled to an Agilent 5977B with extractor EI source (Agilent Technologies). Metabolites of interest were measured in selected ion monitoring mode. The Metabolite Detector software was used for the data analysis with the following settings: peak threshold, 5; minimum peak height, 5; bins per scan, 10; deconvolution width, 5 scans; no baseline adjustment; required peaks, 2; and no minimum required peak intensity. Retention index was calibrated based on the retention time. An in-house mass spectral library was used for compound identification. Mass isotopomer distributions were calculated by MetaboliteDetector's MID wizard. Fractional contribution of glutamine-, glucose-, and palmitate-derived carbon to total metabolite carbon was calculated by dividing the sum of the abundance of all isotopologs (except M0) by the total number of carbons in the respective metabolite.
Statistical Analysis
Single-Cell RNA-Seq Data Analysis
Sequencing data were demultiplexed using Cell Ranger software (version 2.0.2) (10x Genomics), and FASTQ files were generated. Reads were aligned to the UCSC mm10 reference genome (GRCm37) using Cell Ranger followed by quantification of gene expressions and generation of a gene-barcode matrix. Individual datasets were aggregated using the Cellranger aggr command and further analyzed using the R package Seurat (version 3.1.4) (https://cran.r-project.org/package=Seurat). The data were subjected to library-size normalization and log transformation, and the 3,000 most variable genes (based on variance-stabilizing transformation) in the dataset were used for downstream analysis. Principal component analysis (PCA) was used to reduce the dimensionality of the original matrix, and 10 principal components were used to calculate the Uniform Manifold Approximation and Projection (UMAP) and clusters. Genes that were differentially expressed using the FindMarkers function (default parameters) in Seurat and genes with p values <0.01 were considered as differentially expressed genes.
The raw expression matrix was subset by cluster annotation (classical monocytes, immature myeloid cells, and neutrophils) and normalized by SCTransform. The 3,000 most variable genes (as above) were chosen for downstream analysis. Data were scaled, and a PCA was calculated, of which the first 30 components were used for UMAP and clustering. Mutual nearest neighbor batch correction was performed on the low-dimensional representation (PCA) as recommended in the batchelor vignette by Aaron Lun (https://bioconductor.org/packages/release/bioc/vignettes/batchelor/inst/doc/correction.html). Pseudotemporal ordering of single cells was performed using monocle3 using the normalized data (preprocess_cds params: norm_method = “none,” num_dim = 15) including mutual nearest neighbor batch correction (alignment group = sample). Cell cycle assignment was performed using the Seurat function CellCycleScoring using 20 bins and the genes previously reported by Kowalczyk et al. [20]. Over-representation of gene ontology (GO) categories was calculated using the R package clusterProfiler (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html). Visualizations were produced with the R package ggplot2 (https://cran.r-project.org/package=ggplot2).
Other Data Analysis
Comparisons between groups were made using a parametric ANOVA test with the Tukey posttest or a 2-way ANOVA test. p values <0.05 were considered significant. Heatmap of metabolite concentration was generated with R package “pheatmap” with a p value cutoff of 0.05, ANOVA, and z-score normalization. Results are presented as mean values ± SD of a minimum of 3 replicates, and all experiments were repeated at least 3 times.
Results
MDSCs Originate from Both Bone Marrow and Extramedullary Sites in S. aureus Chronic Infection
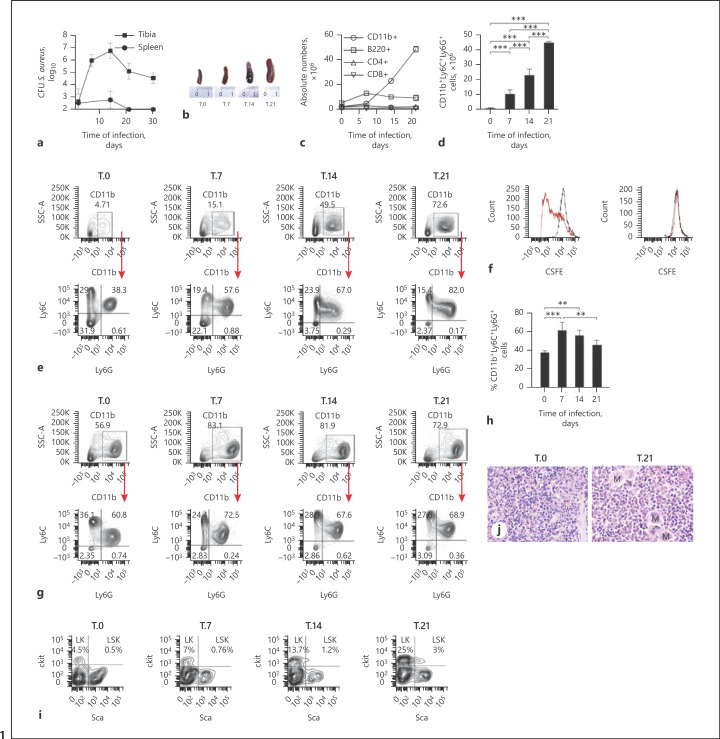
We used a previously described experimental model of S. aureus chronic infection [21] to investigate the origin and physiology of infection-driven MDSCs. C57BL/6 were infected intravenously with 106 CFU of S. aureus strain 6850, and bacterial loads were determined in the tibia and spleen at progressing times after bacterial inoculation. Consistent with previous observations [21], S. aureus was detectable in the tibia of infected mice for up to 30 days, but it was under detection levels in the spleen from day 20 onward (Fig. 1a). Infected mice developed pronounced splenomegaly with the progression of infection (Fig. 1b), which was largely due to a disproportionate accumulation of CD11b+ cells (Fig. 1c) expressing the markers Ly6C and Ly6G (Fig. 1d, e; online suppl. Fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000519306), typical of murine MDSCs [4]. Since the ability to suppress T-cell responses is the hallmark of MDSCs [4], we then assessed the capacity of the Ly6C+Ly6G+ cells accumulating in the spleen of infected mice to inhibit T-cell proliferation. For this purpose, mouse CD4+ T cells isolated from the spleen of uninfected C57BL/6 mice and labeled with CSFE were stimulated with anti-CD3 and anti-CD28 antibodies and incubated in the presence or absence of Ly6C+Ly6G+ cells isolated from the spleen of S. aureus-infected mice at day 21 of infection. On day 3 of culture, proliferation of CD4+ T cells was determined by flow cytometry. As shown in Figure 1f, Ly6C+Ly6G+ potently suppressed proliferation of CD4+ T cells and therefore fulfilled the functional criteria for MDSCs. Furthermore, secretion of cytokines such as IL-2 and IFN-γ by anti-CD3/anti-CD28-stimulated CD4+ T cells was significantly decreased in the presence of MDSCs (online suppl. Fig. 2).
Fig. 1.
MDSCs originate from both the bone marrow and extramedullary sites in S. aureus chronic infection. a Bacterial load in the tibia and spleen of mice after intravenous inoculation with 106 CFU of S. aureus 6850. b Photographs showing splenomegaly in S. aureus-infected mice. c Absolute numbers of CD11b+, B cells (B220+), and CD4+ and CD8+ T cells in the spleen of S. aureus-infected mice at progressing times after bacterial inoculation. d Absolute numbers of CD11b+Ly6C+Ly6G+ in the spleen of S. aureus-infected mice. e Flow cytometry analysis showing the frequency of CD11b+ cells (upper panels) and the frequency of Ly6C+Ly6G+ cells within the CD11b+ population (lower panels) in the spleen of S. aureus-infected mice. Gating strategies are shown in online suppl. Figure 5. f Proliferation of CD4+ T cells unstimulated (black line histograms) or stimulated with anti-CD3ε/anti-CD28 antibodies (red line histograms) for 72 h in the absence (left panel) or presence (right panel) of Ly6C+Ly6G+ cells isolated from the spleen of S. aureus-infected mice at day 21 of infection at a 1:1 ratio. g Flow cytometry analysis showing the frequency of CD11b+ cells (upper panels) and the frequency of Ly6C+Ly6G+ cells within the CD11b+ population (lower panels) in the bone marrow of S. aureus-infected mice. h Percentage of CD11b+Ly6C+Ly6G+ cells in the bone marrow of S. aureus-infected mice. i Representative flow cytometry analysis showing LK and LSK in the spleen of uninfected (T.0) or S. aureus-infected mice at progressing times of infection. j Representative hematoxylin and eosin-stained histological sections of formalin-fixed and paraffin-embedded spleen sections from an uninfected (left) and from a S. aureus-infected (right) mouse at day 21 of infection (M indicates megakaryocytes). Original magnification ×40. Results are presented as the mean ± SD of biological replicates (N = 5 for a and N = 4 for e–h) and are representative of 3 independent experiments. Statistical significance: **p < 0.01; ***p < 0.001. LK, Lin−IL-7Rα−c-Kit+Sca-1− lineage-committed progenitors; LSK, Lin−IL-7Rα−Sca-1+c-Kit+ myeloid progenitors.
We also demonstrated that the expansion of MDSCs during S. aureus chronic infection is not bacterial strain-specific since similar splenomegaly and accumulation of MDSCs with inhibitory effects on T-cell responses were observed in mice intravenously infected with S. aureus strain SH1000, a strain that causes chronic renal infection in mice [22] (online suppl. Fig. 3).
To investigate the origin of the MDSCs arising during S. aureus infection, we first focused on the bone marrow since this is the primary site where myeloid cells are produced. Flow cytometry analysis of the bone marrow isolated from S. aureus-infected mice at progressing times of infection showed a significant increase in the percentage of CD11b+ cells predominantly expressing Ly6C and Ly6G at day 7 that gradually decreased at later times (Fig. 1g, h; online suppl. Fig. 4). Because the kinetic of CD11b+Ly6C+Ly6G+ cells in the bone marrow (Fig. 1g, h; online suppl. Fig. 4) did not match the progressive increase of these cells observed in the spleen during the course of infection (Fig. 1d, e; online suppl. Fig. 1), we speculated that in addition to the bone marrow, MDSCs may also originate from other sites. In this regard, it has been reported that MDSCs can originate at extramedullary sites such as the spleen and liver during chronic inflammatory conditions as a consequence of extramedullary hematopoiesis [5]. To investigate if extramedullary hematopoiesis is occurring at peripheral sites during S. aureus infection, we determined the percentage of Lin−IL-7Rα−c-Kit+Sca-1− lineage-committed progenitors (LK) and of Lin−IL-7Rα−Sca-1+c-Kit+ myeloid progenitors (LSK) in the spleen of infected mice at progressing times after bacterial inoculation. A time-dependent increase in the frequency of LK and of LSK was observed in the spleen of S. aureus-infected mice (Fig. 1i). Histological examination of the spleen tissue taken from S. aureus-infected mice at day 21 of infection showed the red pulp markedly expanded by numerous hematopoietic cells including erythroid and myeloid precursors as well as megakaryocytes, further confirming the occurrence of extramedullary hematopoiesis (Fig. 1j). Together, these results indicate that both the bone marrow and extramedullary hematopoiesis at peripheral sites may contribute to the expansion of MDSCs observed during S. aureus chronic infection.
High-Resolution Mapping of Infection-Driven MDSCs Determined by scRNA-Seq
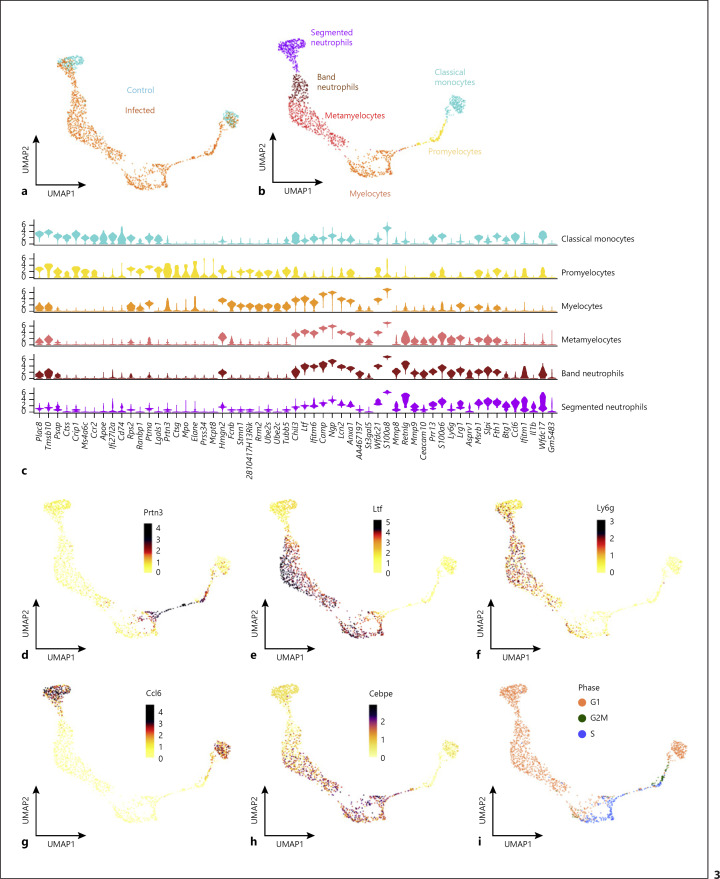
To capture the phenotypic variation among MDSCs present in the spleen of S. aureus-infected mice at a high resolution, we performed scRNA-seq on sorted CD11b+ cells at day 21 of infection. CD11b+ cells isolated from the spleen of uninfected control mice were included to determine the changes in cell composition specifically induced by infection. The scRNA-seq data acquired using the droplet-based 10x Genomics technology from both conditions were combined (1,897 cells from control and 1,497 cells from infected), and a 2-dimensional representation of the single cell transcriptomes was obtained using a UMAP (Fig. 2a). A total of 9 different cell clusters were identified according to the expression of known marker genes, including NK cells (Nkg7, Klrb1c, Klre1, Klrk1, Klra7-9, and Klrd1), B cells (Cd79a, Cd79b, Cd19, and Cd74), dendritic cells (H2-Ab1, H2-Eb1, H2-Aa, Cd209a, and H2-DMb1), classical- (Ly6c2, Ccl9, Ccr2, and Cd68) and nonclassical (Fabp4, Cx3cr1, and Csf1r) monocytes, plasma B cells (Jchain and Sdc1), T cells (Cd3d), neutrophils (S100a8, Ccl6, Il1b, Ly6g, and Wfdc21), and a cluster of immature myeloid cells expressing markers along the granulocytic differentiation axis (Ly6c2, Ly6g, Chil3, Camp, Ltf, S100a8, and Wfdc21) that we classified as MDSCs (Fig. 2b, c; online suppl. Table 1). The proportions of cell types between the 2 conditions are shown in Figure 2d. Notably, the majority of CD11b+ cells from infected mice could be classified as immature myeloid cells, while no such population was present in the control sample.
Fig. 2.
High-resolution map of infection-driven MDSCs determined by scRNA-seq analysis. a UMAP embedding of 1,497 transcriptomes of CD11b+ cells isolated from the spleen of S. aureus-infected mice at day 21 of infection (infected) and 1,897 transcriptomes of CD11b+ cells isolated from uninfected mice (control). b UMAP embedding (as a) showing clusters of cells annotated by cell type. c Dot plot of marker genes underlying the cell type annotation. As indicated by the legend, the size of each dot reflects the proportion of cells of each type expressing the marker gene, and the color intensity reflects the mean expression of each marker gene across all cells in the respective cluster. d Sankey plot showing the proportion of cell types between control and infection. UMAP, Uniform Manifold Approximation and Projection.
To investigate the full extent of heterogeneity of MDSCs, we extracted all transcriptomes annotated as classical monocytes, immature myeloid cells, and neutrophils from the combined dataset and reanalyzed this subset. Conceptionally, the classical monocytes and neutrophils, present in both conditions, represent the typical myeloid cell populations under homeostatic conditions (compare Fig. 3a, b). In infected mice, however, a continuous spectrum of cells could be observed between these 2 populations representing the different stages of granulocyte differentiation as shown in Figure 3b.
Fig. 3.
Single-cell RNA-seq identifies cell populations within infection-driven MDSCs covering the full spectrum of granulocyte differentiation. a UMAP embedding of 1,687 single-cell transcriptomes from the cell clusters classical monocyte, immature myeloid cells, and neutrophils extracted from the combined dataset shown in Figure 2b. b UMAP embedding (as a) showing clusters of cells annotated by cell type. c Violin plots showing the expression of selected marker genes of the different cell clusters depicted in b. Expression in each cell population is shown along with the probability density of gene expression symbolized by the shape of the plot. d–h UMAP embeddings (as a) showing the normalized expression of Prtn3 (d), Ltf (e), Ly6g (f), Ccl6 (g), and Cebpe (h) genes. i UMAP embedding showing the cell cycle phase. UMAP, Uniform Manifold Approximation and Projection.
Neutrophils contain 4 types of granules including primary (azurophilic), secondary (specific), tertiary (gelatinase), and ficolin-1-rich granules. These granules are produced stepwise during the different stages of maturation that start with promyelocytes followed by myelocytes, metamyelocytes, band cells, and end with terminally differentiated segmented neutrophils [23]. This process has been described as a targeting-by-timing model to explain the differences in protein contents among neutrophil granule subsets [23, 24, 25, 26]. Accordingly, we used the expression levels of the genes encoding the different granule proteins to classify the spectrum of cell populations identified by scRNA-seq within the MDSCs into specific neutrophil differentiation categories. Azurophilic granule proteins including myeloperoxidase (encoded by Mpo), elastase (encoded by Elane), cathepsin G (encoded by Ctsg), and proteinase 3 (encoded by Prtn3) are produced only at the promyelocyte stage (Fig. 3c, d; online suppl. Table 2). Myelocytes were identified by the high expression of genes encoding secondary granule proteins such as lactoferrin (Ltf), cathelicidin (Camp), and neutrophil gelatinase-associated lipocalin (Lcn2) as well as by the expression of the gene encoding ficolin-1 (Fcnb), which originates during the transition from myelocytes to metamyelocytes (Fig. 3c, e; online suppl. Table 2). Metamyelocytes could be identified based on the high expression of the gene encoding the above-mentioned secondary granule proteins (Ltf, Camp, Lcn2, and Fcnb) and by the increased expression of the gene encoding Ly6G (Ly6g) (Fig. 3c-f; online suppl. Table 2). The expression of genes encoding tertiary granule proteins such as metalloproteinase 8 (Mmp8) and metalloproteinase 9 (Mmp9) identified band neutrophils (Fig. 3c; online suppl. Table 2). Last, terminally differentiated segmented neutrophils were identified based on the high expression of genes encoding markers such as colony-stimulating factor 3 receptor (Csf3r), IL1-β (Il1b), and CCL6 (Ccl6) (Fig. 3c, g; online suppl. Table 2). The expression pattern of Cebpe, which encodes the transcription factor CCAAT/enhancer binding protein-ε (C/EBP-ε) that is predominantly expressed during the myelocyte and metamyelocyte differentiation stages [27], confirmed the classification performed based on the expression of genes encoding granule proteins (Fig. 3h; online suppl. Table 2).
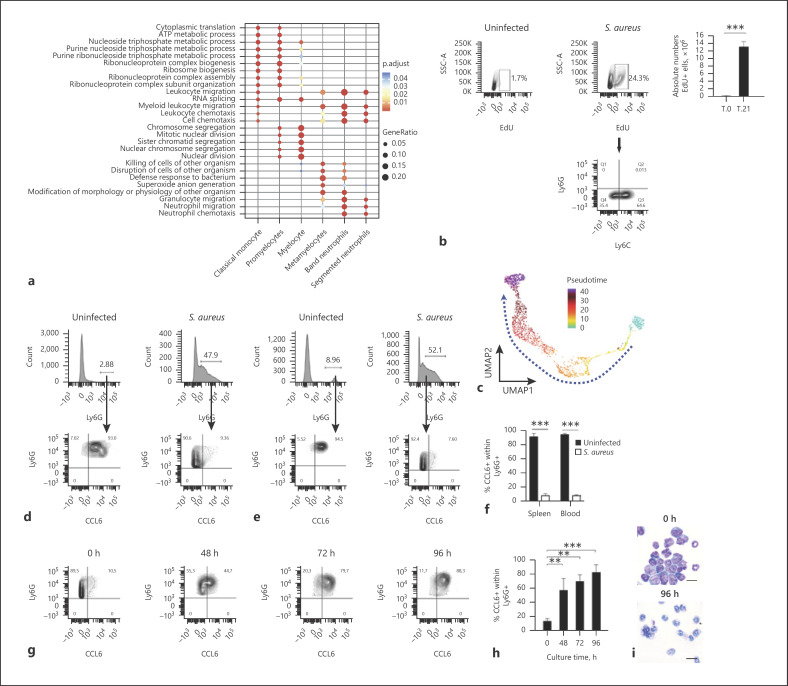
Mature neutrophils are mitotically inactive with cell cycle arrest occurring during the myelocyte to metamyelocyte transition [25]. To substantiate this in our experimental setting, we performed cell cycle analysis in the scRNA-seq data. The results indicated that both promyelocytes and myelocytes were actively proliferating as they were in phases S (DNA synthesis) and G2/M (cell division) of the cell cycle (Fig. 3i). On the other hand, metamyelocytes, band neutrophils, and segmented neutrophils were all in the postmitotic G1 phase (growth phase, Fig. 3i). Consistent with these results, gene ontology analysis identified the cell cycle to be an overrepresented functional category in promyelocytes and myelocytes, whereas functional categories associated with cell migration and host defense were overrepresented in the more mature populations such as band and segmented neutrophils (Fig. 4a). In addition, we performed functional analysis to determine the presence of actively proliferating cells in the spleen of S. aureus-infected mice in vivo using EdU, a thymidine analog that is incorporated into proliferating cells during DNA synthesis. A significantly higher number of proliferating cells were detected in the spleen of S. aureus-infected mice (day 21 of infection) in comparison to uninfected control mice (Fig. 4b, upper panels). More than 60% of the actively proliferating cells (EdU+) were Ly6C+/Ly6G− (Fig. 4b, lower panel) and most probably represented promyelocyte and myelocyte cell populations.
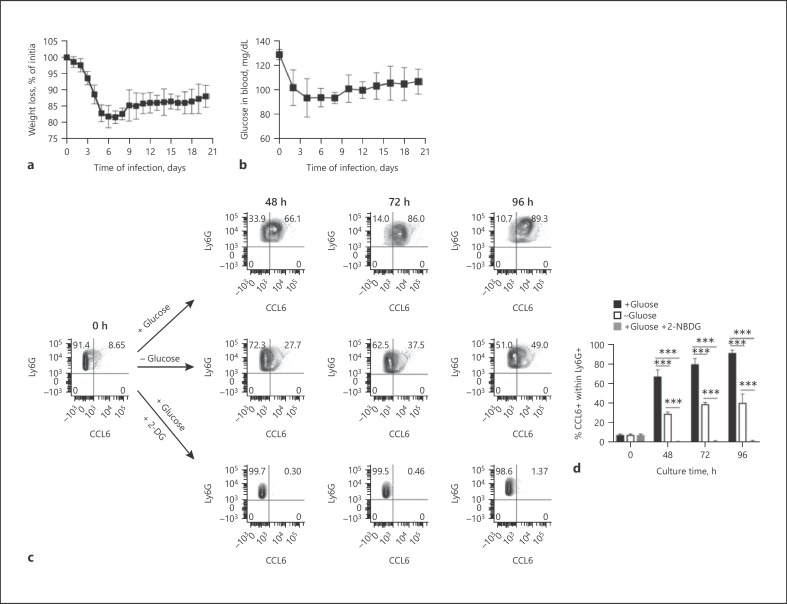
Fig. 4.
Reconstruction of trajectories and transition states of infection-driven MDSCs. a Most enriched GO categories of the upregulated (ranked by log-fold change) genes for each cell cluster depicted in Figure 3b. b Representative flow cytometry contour plots showing the percentage of EdU+ cells gated in the total spleen cell population of uninfected mice (left panel) and S. aureus-infected mice at day 21 of infection (middle panel). The absolute numbers of EdU+ cells in the spleen of uninfected (T.0) and S. aureus-infected (T.21) mice are shown in the right panel. The expression of Ly6G and Ly6C within the EdU+ cell population in the spleen of S. aureus-infected mice is shown in the contour plot depicted in the lower panel. The gating strategy is described in online suppl. Figure 6. c UMAP embedding showing granulocyte differentiation as a continuous spectrum of pseudotime. The dashed line indicates the progression toward terminal differentiation. d, e Flow cytometry analysis of CCL6 expression within the Ly6G+ population in the spleen (d) and blood (e) of uninfected (left panels) or S. aureus-infected (right panels) mice at day 21 of infection. f Percentage of CCL6+ cells within Ly6G+ cell population in the spleen and blood on uninfected (black bars) or S. aureus-infected (white bars) mice. g Flow cytometry contour plots showing the expression kinetic of CCL6 within Ly6G+ cells isolated from the spleen of S. aureus-infected mice at day 21 of infection and cultured in vitro for 96 h. The gating strategy is depicted in online suppl. Figure 8. h Percentage of CCL6+ cells within the Ly6G+ cell population at progressive times during in vitro culture. i May-Grünwald-Giemsa staining of cytospin preparations from Ly6G+ spleen cells isolated from S. aureus-infected mice at day 21 of infection before (upper panel) and 96 h after (lower panel) in vitro culture. Scale bars represent 200 μm. Results are presented as the mean ± SD of 3 biological replicates (N = 3) and are representative of 3 independent experiments. Statistical significance: **p < 0.01; ***p < 0.001. UMAP, Uniform Manifold Approximation and Projection; GO, gene ontology; EdU, 5-ethynyl-2′-deoxyuridine.
To further validate the hierarchy between the different myeloid cell populations within the MDSC cluster identified by the scRNA-seq data, we performed trajectory analysis based on pseudotime, where cells represent distinct stages in a continuous developmental process. This enables the association of specific cell types with the initial, intermediate, and terminal states of the trajectory [28]. The pseudotime analysis shown in Figure 4c recapitulated the trajectory of cell differentiation from promyelocytes (initial) to terminally differentiated segmented neutrophils (final) including several intermediate developmental states comprising myelocytes, metamyelocytes, and band neutrophils.
Distinguishing between immature granulocytic precursors and mature segmented neutrophils has been very difficult, and no phenotypic marker has been identified so far that enables precise separation of these populations. The results of the scRNA-seq analysis performed in this study have revealed that the expression of both Ly6G and CCL6 markers may be suitable to separate mature neutrophils (Ly6G+CCL6+) from immature MDSC precursors (Ly6G+CCL6−). This was corroborated by flow cytometry analysis showing that whereas approximately 90% of neutrophils in the spleen (Fig. 4d, f) and blood (Fig. 4e, f) of uninfected mice were mature neutrophils (Ly6G+CCL6+), <10% of Ly6G+ cells expressed CCL6 in the spleen (Fig. 4d, f) and blood (Fig. 4e, f) of S. aureus-infected mice.
In the cancer setting, it has been reported that MDSCs are not irreversibly arrested in an immature stage and could terminally differentiate after being removed from the tumor environment and cultured under in vitro conditions [29, 30, 31]. To investigate if this was also the case for infection-driven MDSCs, we determined the capacity of MDSCs isolated from the spleen of S. aureus-infected mice at day 21 of infection to undergo terminal maturation upon in vitro culture conditions. As the surface expression of Ly6G and the intracellular expression of CCL6 were revealed by the scRNA-seq analysis as markers of mature neutrophils, we monitored MDSC maturation by measuring the level of expression of these markers at increasing times of in vitro culture. Although cell viability slowly decreased with time, over 70% of Ly6G+cells were still viable after 96 h of in vitro culture (online suppl. Fig. 7a, b). Flow cytometry analysis of Ly6G+ at different times of in vitro culture showed a time-dependent gradual increase in the percentage of Ly6G+CCL6+ (Fig. 4g, h). After 96 h of in vitro culture, >80% of Ly6G+ cells expressed CCL6, when only 10% of the Ly6G+ expressed CCL6 prior to in vitro culture (0 h) (Fig. 4g, h). Morphological changes were also observed in in vitro-cultured MDSCs, which showed a transition from predominant immature myeloid cells including cells exhibiting round nuclei typical of promyelocytes, kidney-shaped nuclei typical of myelocytes and metamyelocytes, and band-like-shaped nuclei typical of band neutrophils prior to in vitro culture (Fig. 4i, upper panel) to cells with segmented nuclei morphology typical of mature neutrophils after 96 h of in vitro culture (Fig. 4i, lower panel). Furthermore, the in vitro-cultured Ly6C+Ly6G+ cells were capable of phagocytizing and killing internalized S. aureus (online suppl. Fig. 9). Together, these results indicate that S. aureus infection-driven MDSCs retained their capacity to terminally differentiate into mature myeloid cells and can undergo maturation after being removed from the spleen environment.
Infection-Driven Immature MDSCs Rely on Aerobic Glycolysis to Complete Their Maturation Process
As metabolism has been shown to influence immune cell differentiation and function [32], a better understanding of the metabolic pathways used by MDSCs to support both their energetic and biosynthetic demands could provide important information about their difficulties for undergoing terminal differentiation.
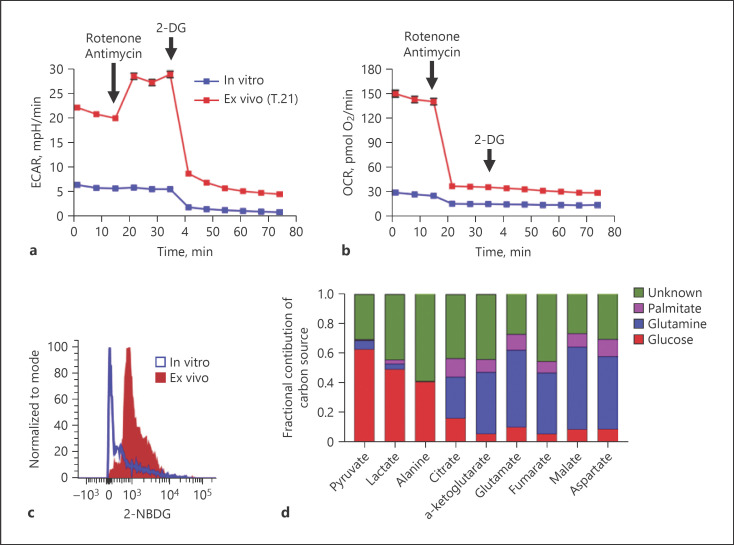
Therefore, we analyzed ECAR as a surrogate for glycolytic rate and OCR as an indicator of mitochondrial oxidative phosphorylation in immature Ly6C+Ly6G+ cells directly isolated from the spleen of S. aureus-infected mice at day 21 of infection (ex vivo) and in Ly6C+Ly6G+ cells after maturation in in vitro culture for 96 h (in vitro) using a Seahorse XF biochemical analyzer [33]. Ex vivo immature Ly6C+Ly6G+ cells showed increased basal ECAR (Fig. 5a) and OCR (Fig. 5b) as compared to in vitro-cultured mature Ly6C+Ly6G+ cells, indicating that immature Ly6C+Ly6G+ cells had increased energetic demands compared with mature Ly6C+Ly6G+ cells. Injection of a mixture of complex I inhibitor rotenone and complex III inhibitor antimycin A blocked mitochondrial respiration in both ex vivo immature and in vitro-cultured mature Ly6C+Ly6G+ cells, as evidenced by reduced OCR values (Fig. 5b). The decrease in OCR in ex vivo immature Ly6C+Ly6G+ cells was accompanied by an increase in ECAR (Fig. 5a), indicating that glycolysis is induced after inhibition of the electron transport chain to compensate ATP production and to meet their energy demand. Subsequent injection of 2-DG, a glucose analog that inhibits hexokinase, the first enzyme in the glycolysis pathway, resulted in substantial reduction of ECAR below the basal level in both ex vivo immature and in vitro-cultured mature Ly6C+Ly6G+ cells (Fig. 5a), confirming thus that acidification of the medium after inhibition of mitochondrial respiration was driven by glycolysis. These results imply that infection-driven immature MDSCs used both aerobic glycolysis and oxidative phosphorylation to support their bioenergetic demands.
Fig. 5.
Infection-driven MDSCs rely heavily on aerobic glycolysis and glucose uptake to meet their bioenergetic demands. a, b Extracellular flux measurements of ECAR (a) and OCR (b) in splenic Ly6C+Ly6G+ cells isolated from S. aureus-infected mice (day 21 of infection) prior to (ex vivo, red symbols) and after in vitro culture for 96 h (in vitro, blue symbols) determined by a Seahorse XF96 analyzer. c Representative histogram showing fluorescence 2-NBDG uptake by Ly6C+Ly6G+ cells isolated from S. aureus-infected mice (day 21 of infection) prior to (ex vivo, red histogram) and after in vitro culture for 96 h (in vitro, blue histogram). d Fractional contribution of different carbon sources to the total carbon of cellular metabolites in Ly6C+Ly6G+ cells isolated from the spleen of S. aureus-infected mice (day 21 of infection) determined by isotopic tracing with [U-13C6]-glucose, [U-13C5]-glutamine, or [U-13C16]-palmitate and GC-MS measurement. ECAR, extracellular acidification rate; OCR, oxygen consumption rate; 2-DG, 2-deoxy-D-glucose; 2-NBDG, 2-(N-[7-nitrobenz-2-oxa-1,3-diazol-4-yl] amino)-2-deoxyglucose.
In accordance with their high glycolytic activity, ex vivo immature Ly6C+Ly6G+ cells consumed greater amounts of glucose than in vitro-cultured mature Ly6C+Ly6G+ cells as determined by flow cytometry using the fluorescent D-glucose analog 2-(N-[7-nitrobenz-2-oxa-1,3-diazol-4-yl] amino)-2-deoxy-D-glucose (Fig. 5c). As ex vivo immature Ly6C+Ly6G+ MDSCs also utilized oxidative phosphorylation to fulfill their bioenergetic requirements, we also determined which carbon sources were used by these cells to support oxidative metabolism. For this purpose, ex vivo immature Ly6C+Ly6G+ cells were incubated with [U-13C6]-glucose, [U-13C5]-glutamine, or [U-13C16]-palmitate, and the labeling pattern of selected metabolites was determined by GC-MS measurement. As expected, glucose-derived carbon was incorporated not only into pyruvate but also into lactate, further corroborating the use of aerobic glycolysis by S. aureus infection-driven immature MDSCs (Fig. 5d). The flux of glutamine-derived carbon into TCA cycle intermediates showed that approximately 50% of carbon in the TCA cycle is derived from glutamine (Fig. 5d). This may indicate that due to the excessive conversion of glucose into lactate, MDSCs used anaplerosis of glutamine to replenish TCA cycle intermediates. The results depicted in Figure 5d also show that glucose and palmitate fueled the TCA cycle with acetyl-CoA at comparable levels.
Infection-Driven Immature MDSCs Are Reliant on Glucose Availability for Terminal Differentiation
We next investigated the reason why MDSCs failed to complete their maturation program and accumulated in the spleen of infected mice in an immature stage of differentiation. The metabolic analysis performed above indicated that MDSCs exhibited high glycolytic activity and high rate of glucose consumption that may support the elevated biosynthetic requirements associated with the maturation process. However, glucose may become rapidly depleted in the spleen microenvironment due to its rapid consumption by the increased proportion of MDSCs accumulating in this organ. Furthermore, glucose supply during infection may be insufficient as a consequence of reduced food intake by infected mice. This was particularly evident in our study since S. aureus-infected mice exhibited progressive weight loss during the course of infection (Fig. 6a) and exhibited reduced concentrations of glucose in blood (Fig. 6b). Based on these observations, we postulated that limited glucose availability during infection may pose a bottleneck for MDSCs to undergo complete maturation. To substantiate this assumption, we investigated if the level of glucose availability influenced the maturation status of MDSCs. For this purpose, we determined the capacity of immature MDSCs isolated from the spleen of S. aureus-infected mice to undergo terminal maturation under in vitro culture conditions in the presence or absence of glucose or after inhibition of glycolysis by measuring the level of expression of surface Ly6G and intracellular CCL6. The results show that approximately 90% of MDSCs underwent terminal maturation at 96 h of culture in the presence of glucose, but only 40% underwent terminal maturation in cultures where glucose was removed from the culture medium (Fig. 6c, d). Terminal differentiation of MDSCs was completely suppressed when the inhibitor of glycolysis 2-DG was added to the cultures (Fig. 6c, d). Cell viability was significantly lower in the absence of glucose or after inhibition of glycolysis with 2-DG than in cells cultured in the presence of glucose (online suppl. Fig. 10a).
Fig. 6.
Infection-driven MDSCs are reliant on glucose availability for terminal differentiation. a Percentage of weight loss of S. aureus-infected mice during the course of infection. b Glucose levels in the blood of S. aureus-infected mice during the course of infection. c Representative flow cytometry contour plots showing the expression of CCL6 within Ly6G+ cells isolated from the spleen of S. aureus-infected mice at day 21 of infection and cultured in vitro either in the presence or absence of glucose or after inhibition of glycolysis by addition of 2-DG. d Percentage of CCL6+ cells within the Ly6G+ cell population at progressive times during in vitro culture either in the presence (black bars) or absence (white bars) of glucose or after inhibition of glycolysis with 2-DG (gray bars). Results are presented as the mean ± SD of biological replicates (N = 5 for a, b and N = 3 for c, d) and are representative of 3 independent experiments. Statistical significance: ***p < 0.001. 2-DG, 2-deoxy-D-glucose.
The impact of glucose availability on MDSC maturation was further confirmed by a trend toward a reduction in the terminal maturation of MDSCs observed upon exposure to decreased glucose concentrations (online suppl. Fig. 10b, c). Furthermore, only MDSCs cultured in vitro for 96 h in the presence of 2 mg/mL of glucose exhibited significantly lower capacity to inhibit T-cell proliferation than immature MDSCs prior to culture (online suppl. Fig. 10d).
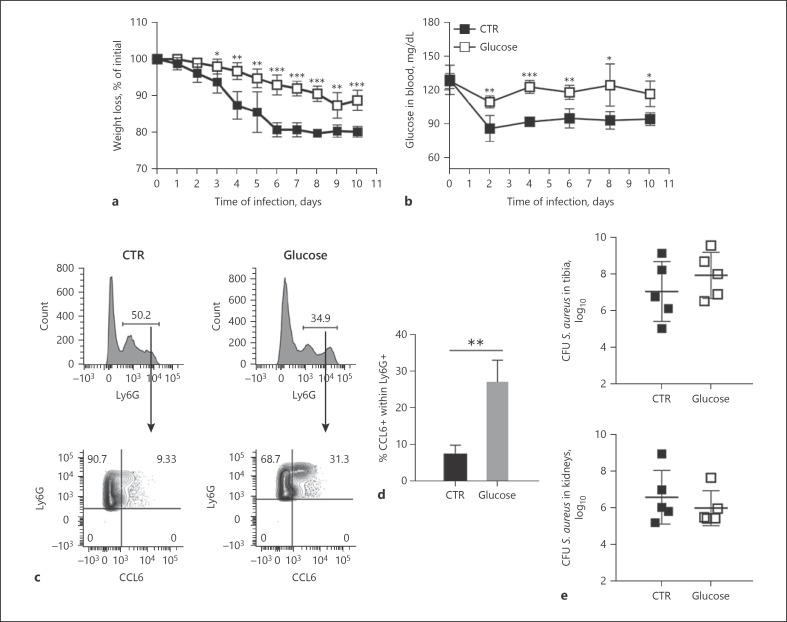
Based on these observations, we next investigated the effect of increasing glucose availability in vivo by supplementing S. aureus-infected mice with 10% glucose in drinking water for 10 days on the maturation of splenic MDSCs. Glucose-treated mice exhibited significantly less body weight loss (Fig. 7a) and higher levels of glucose in blood (Fig. 7b) than untreated mice. Importantly, glucose-treated mice exhibited a significantly higher number of mature neutrophils (Ly6G+CCL6+) in the spleen than the untreated group (Fig. 7c, d). However, glucose supplementation did not affect the bacterial loads in infected organs (Fig. 7e).
Fig. 7.
Supplying S. aureus-infected mice with glucose accelerated differentiation of MDSCs in vivo. Percent weight loss (a) and glucose levels in the blood (b) of S. aureus-infected mice fed with glucose-supplemented (white symbols) or nonsupplemented (black symbols) drinking water for 10 days. c Representative flow cytometry analysis showing the expression of CCL6 within the Ly6G+ cell population in the spleen of S. aureus-infected mice (day 10 of infection) fed with glucose-supplemented (right panels) or nonsupplemented (left panels) drinking water. d Percentage of CCL6+ cells within the Ly6G+ cell population in the spleen of S. aureus-infected mice (day 10 of infection) that received glucose-supplemented (gray bar) or nonsupplemented (black bar) drinking water. e Bacterial load in the tibia (upper panel) and kidneys (lower panel) of S. aureus-infected mice (day 10 of infection) that received glucose-supplemented (white symbols) or nonsupplemented (black symbols) drinking water. Results are presented as the mean ± SD of collated data from 3 independent experiments (N = 9). Statistical significance: **p < 0.01; ***p < 0.001.
Discussion
In this study, we integrated scRNA-seq analysis and functional metabolic profiling to gain a deeper understanding of the generation and physiology of MDSCs in the context of S. aureus chronic infection. The results of the scRNA-seq analysis emphasize the vast heterogeneity and functional diversity of infection-driven MDSCs, which comprise a continuous spectrum of cell populations representing transitions between different states of granulocyte differentiation, ranging from promyelocytes to mature segmented neutrophils. In mice, the phenotypic distinction between mature neutrophils and immature progenitors has been difficult, and no phenotypic marker has been identified so far that enables to precisely separate these populations. The scRNA-seq analysis performed in our study has identified surface expression of Ly6G and intracellular expression of CCL6 as phenotypic markers that enable to distinguish mature neutrophils (Ly6G+CCL6+) from immature granulocytes precursors (Ly6G+CCL6−) by flow cytometry. However, CCL6 has the drawback to be intracellular and its detection requires cell fixation and therefore does not allow recovery of live cells for subsequent functional studies. To date, relatively little information is available about the function and expression of CCL6. In mice, CCL6 is expressed in cells of granulocyte and macrophage lineages and is highly induced upon stimulation with GM-CSF [34]. Other studies have reported a role for this chemokine in inflammation and tissue remodeling [18]. The human homolog of CCL6 has not yet been identified, an issue that deserves further attention in future studies.
We found that infection-driven MDSCs originate from both the bone marrow, most probably as a consequence of emergency granulopoiesis, and in situ within the spleen from extramedullary hematopoiesis. Emergency granulopoiesis induced by infection and the concomitant release of immature myeloid cells in the circulation seem to be a mechanism triggered to restore the neutrophil pool that is rapidly depleted from peripheral blood due to extravasation from the bloodstream into the sites of infection [35]. We also investigated the reason why infection-driven MDSCs fail to undergo terminal maturation and accumulate in an early stage of differentiation. We found that, similar to other pathological conditions [29, 30, 31], MDSCs in chronically infected mice are not irreversibly arrested in an immature stage and still retain their capacity to terminally differentiate into mature myeloid cells under in vitro culture conditions. Since metabolism plays an important role in immune cell differentiation and function [32], we explored a possible connection between the metabolic demands of MDSCs and their difficulties for undergoing terminal differentiation. The metabolic flux and isotope tracing analysis performed in our study indicate that infection-driven MDSCs use both aerobic glycolysis and oxidative phosphorylation for ATP production. The benefits of aerobic glycolysis for MDSCs may be both bioenergetics and biosynthesis. During glycolysis, 1 molecule of glucose is converted into 2 molecules of pyruvate with the concomitant generation of 2 molecules of ATP. Generally, pyruvate enters the mitochondria where it is converted into acetyl-CoA, which enters the TCA cycle. In certain circumstances, as those observed in our study in the infection-driven MDSCs, a proportion of pyruvate can be also converted into lactate in the cytosol by lactate dehydrogenase with concomitant regeneration of NAD+ from NADH that keeps fueling the glycolytic pathway. Therefore, although the ATP generated per glucose molecule during aerobic glycolysis is rather low, a very high glycolytic flux like that detected in MDSCs from infected mice can produce high levels of ATP. Furthermore, in addition to ATP generation, glycolysis may provide biosynthetic intermediates to support the synthesis of important molecular building blocks required by MDSCs for undergoing differentiation and maturation.
Based on the results of the metabolic analysis, we speculated that MDSCs may rely on high glycolytic activity to complete their maturation process and that glucose limitation in the spleen microenvironment, possibly due to its rapid consumption by MDSCs and/or to a decline in glucose blood concentrations observed in infected mice, could prevent their complete maturation. This assumption proved to be true since MDSCs isolated from infected mice were capable of undergoing terminal differentiation under in vitro conditions when glucose was added to the culture medium, but differentiation was hampered in the absence of glucose or when glycolysis was inhibited. Furthermore, we could show that supplying S. aureus-infected mice with glucose in the drinking water resulted in improved blood glucose levels, ameliorated weight loss, and accelerated differentiation of immature myeloid cells in the spleen. However, the bacterial loads in the organs of infected mice were not affected by glucose supplementation. One possible explanation for this phenomenon could be that a proportion of MDSCs were still present in the glucose-treated mice that could interfere with effective T-cell responses. Furthermore, as glucose is the principal energy source of S. aureus, increasing glucose levels in treated mice could enhance S. aureus pathogenesis as reported by previous studies [36, 37, 38, 39].
In summary, the results of our study have uncovered a link between metabolic alterations induced by infection and the accumulation of MDSCs.
Statement of Ethics
Animal experiments were performed in strict accordance with the German regulations of the Society for Laboratory Animal Science (GV-SOLAS) and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA). All experiments were approved by the ethical board Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany (LAVES; permit Nos. 33.9-42502-04-10/0296 and 33.19-42502-04-19/3307).
Conflict of Interest Statement
The authors declare no competing interests.
Funding Sources
This work was supported in part by internal funding provided by the Helmholtz Centre for Infection Research.
O.D. and A.-E.S. are supported by the Deutsche Forschungsgemeinschaft (DFG) GRK 2157 3D Tissue Models for Studying Microbial Infections by Human Pathogens.
Author Contributions
O.G. and E.M. conceived and designed the study, performed the in vivo experiments, and analyzed the data. O.D., R.G., and A.-E.S. carried out the scRNA-seq procedure and analysis. A.H. and K.H. performed the metabolic analysis. A.B. performed histological evaluation. E.M. wrote the manuscript with input from all authors.
Data Availability Statement
The raw bulk RNA-seq data and raw single-cell RNA-seq data generated during this study are available at the European Nucleotide Archive (ENA) under the accession number: PRJEB38066.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
The authors would like to thank S. Beyer and P. Hagendorff for excellent technical assistance.
References
- 1.Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017 Jan;5((1)):3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018 Feb;19((2)):108–19. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009 Mar;9((3)):162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016 Jul 6;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuenca AG, Delano MJ, Kelly-Scumpia KM, Moreno C, Scumpia PO, Laface DM, et al. A paradoxical role for myeloid-derived suppressor cells in sepsis and trauma. Mol Med. 2011 Mar–Apr;17((3–4)):281–92. doi: 10.2119/molmed.2010.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldmann O, Beineke A, Medina E. Identification of a novel subset of myeloid-derived suppressor cells during chronic staphylococcal infection that resembles immature eosinophils. J Infect Dis. 2017 Dec 12;216((11)):1444–51. doi: 10.1093/infdis/jix494. [DOI] [PubMed] [Google Scholar]
- 7.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012 Mar 22;12((4)):253–68. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medina E, Hartl D. Myeloid-derived suppressor cells in infection: a general overview. J Innate Immun. 2018;10((5–6)):407–13. doi: 10.1159/000489830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heim CE, Vidlak D, Scherr TD, Kozel JA, Holzapfel M, Muirhead DE, et al. Myeloid-derived suppressor cells contribute to Staphylococcus aureus orthopedic biofilm infection. J Immunol. 2014 Apr 15;192((8)):3778–92. doi: 10.4049/jimmunol.1303408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heim CE, Vidlak D, Kielian T. Interleukin-10 production by myeloid-derived suppressor cells contributes to bacterial persistence during Staphylococcus aureus orthopedic biofilm infection. J Leukoc Biol. 2015 Dec;98((6)):1003–13. doi: 10.1189/jlb.4VMA0315-125RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heim CE, Vidlak D, Scherr TD, Hartman CW, Garvin KL, Kielian T. IL-12 promotes myeloid-derived suppressor cell recruitment and bacterial persistence during Staphylococcus aureus orthopedic implant infection. J Immunol. 2015 Apr 15;194((8)):3861–72. doi: 10.4049/jimmunol.1402689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tebartz C, Horst SA, Sparwasser T, Huehn J, Beineke A, Peters G, et al. A major role for myeloid-derived suppressor cells and a minor role for regulatory T cells in immunosuppression during Staphylococcus aureus infection. J Immunol. 2015 Feb 1;194((3)):1100–11. doi: 10.4049/jimmunol.1400196. [DOI] [PubMed] [Google Scholar]
- 13.Heim CE, Vidlak D, Odvody J, Hartman CW, Garvin KL, Kielian T. Human prosthetic joint infections are associated with myeloid-derived suppressor cells (MDSCs): implications for infection persistence. J Orthop Res. 2018 Jun;36((6)):1605–13. doi: 10.1002/jor.23806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heim CE, West SC, Ali H, Kielian T. Heterogeneity of Ly6G(+) Ly6C(+) myeloid-derived suppressor cell infiltrates during Staphylococcus aureus biofilm infection. Infect Immun. 2018 Dec;86((12)):e00684-18. doi: 10.1128/IAI.00684-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015 Jul;28((3)):603–61. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law AMK, Valdes-Mora F, Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for cancer. Cells. 2020 Feb 27;9((3)):561. doi: 10.3390/cells9030561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta. 2011 Jun;1807((6)):726–34. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 18.Coelho AL, Schaller MA, Benjamim CF, Orlofsky AZ, Hogaboam CM, Kunkel SL. The chemokine CCL6 promotes innate immunity via immune cell activation and recruitment. J Immunol. 2007 Oct 15;179((8)):5474–82. doi: 10.4049/jimmunol.179.8.5474. [DOI] [PubMed] [Google Scholar]
- 19.Sapcariu Sean C, Kanashova Tamara, Weindl Daniel, Ghelfi Jenny, Dittmar Gunnar, Hiller Karsten. Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX. 2014;1:74–80. doi: 10.1016/j.mex.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015 Dec;25((12)):1860–72. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horst SA, Hoerr V, Beineke A, Kreis C, Tuchscherr L, Kalinka J, et al. A novel mouse model of Staphylococcus aureus chronic osteomyelitis that closely mimics the human infection: an integrated view of disease pathogenesis. Am J Pathol. 2012 Oct;181((4)):1206–14. doi: 10.1016/j.ajpath.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler C, Goldmann O, Hobeika E, Geffers R, Peters G, Medina E. The dynamics of T cells during persistent Staphylococcus aureus infection: from antigen-reactivity to in vivo anergy. EMBO Mol Med. 2011 Nov;3((11)):652–66. doi: 10.1002/emmm.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowland JB, Borregaard N. Granulopoiesis and granules of human neutrophils. Immunol Rev. 2016 Sep;273((1)):11–28. doi: 10.1111/imr.12440. [DOI] [PubMed] [Google Scholar]
- 24.Cowland JB, Borregaard N. The individual regulation of granule protein mRNA levels during neutrophil maturation explains the heterogeneity of neutrophil granules. J Leukoc Biol. 1999 Dec;66((6)):989–95. doi: 10.1002/jlb.66.6.989. [DOI] [PubMed] [Google Scholar]
- 25.Theilgaard-Monch K, Jacobsen LC, Borup R, Rasmussen T, Bjerregaard MD, Nielsen FC, et al. The transcriptional program of terminal granulocytic differentiation. Blood. 2005 Feb 15;105((4)):1785–96. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 26.Rorvig S, Ostergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 2013 Oct;94((4)):711–21. doi: 10.1189/jlb.1212619. [DOI] [PubMed] [Google Scholar]
- 27.Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood. 2003 Jun 1;101((11)):4322–32. doi: 10.1182/blood-2002-03-0835. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths JA, Scialdone A, Marioni JC. Using single-cell genomics to understand developmental processes and cell fate decisions. Mol Syst Biol. 2018 Apr 16;14((4)):e8046. doi: 10.15252/msb.20178046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bronte V, Apolloni E, Cabrelle A, Ronca R, Serafini P, Zamboni P, et al. Identification of a CD11b(+)/Gr-1(+)/CD31(+) myeloid progenitor capable of activating or suppressing CD8(+) T cells. Blood. 2000 Dec 1;96((12)):3838–46. [PMC free article] [PubMed] [Google Scholar]
- 30.Kusmartsev S, Gabrilovich DI. Inhibition of myeloid cell differentiation in cancer: the role of reactive oxygen species. J Leukoc Biol. 2003 Aug;74((2)):186–96. doi: 10.1189/jlb.0103010. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Pan PY, Gu P, Xu D, Chen SH. Role of immature myeloid Gr-1+ cells in the development of antitumor immunity. Cancer Res. 2004 Feb 1;64((3)):1130–9. doi: 10.1158/0008-5472.can-03-1715. [DOI] [PubMed] [Google Scholar]
- 32.Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell. 2017 May 4;169((4)):570–86. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Nuebel E, Wisidagama DR, Setoguchi K, Hong JS, Van Horn CM, et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat Protoc. 2012 May 10;7((6)):1068–85. doi: 10.1038/nprot.2012.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orlofsky A, Berger MS, Prystowsky MB. Novel expression pattern of a new member of the MIP-1 family of cytokine-like genes. Cell Regul. 1991 May;2((5)):403–12. doi: 10.1091/mbc.2.5.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014 May;14((5)):302–14. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 36.Rich J, Lee JC. The pathogenesis of Staphylococcus aureus infection in the diabetic NOD mouse. Diabetes. 2005 Oct;54((10)):2904–10. doi: 10.2337/diabetes.54.10.2904. [DOI] [PubMed] [Google Scholar]
- 37.Waldrop R, McLaren A, Calara F, McLemore R. Biofilm growth has a threshold response to glucose in vitro. Clin Orthop Relat Res. 2014 Nov;472((11)):3305–10. doi: 10.1007/s11999-014-3538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Equils O, da Costa C, Wible M, Lipsky BA. The effect of diabetes mellitus on outcomes of patients with nosocomial pneumonia caused by methicillin-resistant Staphylococcus aureus: data from a prospective double-blind clinical trial comparing treatment with linezolid versus vancomycin. BMC Infect Dis. 2016 Sep 6;16:476. doi: 10.1186/s12879-016-1779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitko NP, Grosser MR, Khatri D, Lance TR, Richardson AR. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio. 2016 Jun 21;7((3)):e00296-16. doi: 10.1128/mBio.00296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The raw bulk RNA-seq data and raw single-cell RNA-seq data generated during this study are available at the European Nucleotide Archive (ENA) under the accession number: PRJEB38066.