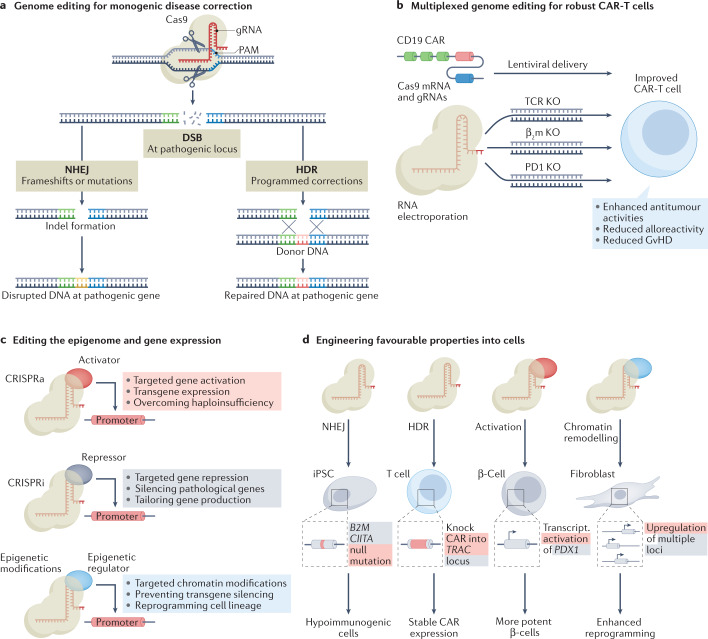
Fig. 2. Leveraging CRISPR–Cas-mediated genome and epigenome editing for improved cell-based therapeutics.
a | Genome editing can be applied to correct monogenic diseases. Double strand breaks (DSBs) resulting from programmed genome editing outcomes generally resolve via non-homologous end joining (NHEJ) or homology-directed repair (HDR) repair mechanisms in human cells. NHEJ typically results in insertions or deletions (indels) near the targeted genomic site, which can be leveraged for programmable endogenous genetic disruption. By contrast, in the presence of a donor DNA template, HDR can permit precision replacement of genomic DNA, including donor templated to correct DNA associated with pathology or to incorporate clinically important transgenic payloads. b | CRISPR–Cas-based genome editing technologies are highly amenable to multiplexing, which can be used to improve cell-based therapeutics, including chimeric antigen receptor T cell (CAR-T) therapies. Multiplexed CRISPR–Cas9-based genome editing (shown here simultaneously targeting the genes encoding human β2-microglobulin (β2m), PD1 and endogenous T cell receptor (TCR)) in combination with a lentivirally delivered CAR can be used to generate CAR-T cells with improved function and safety profiles. c | CRISPR–Cas systems with inactivated nuclease activity do not result in DSBs, but can still precisely target genomic DNA. These CRISPR–Cas-based ‘epigenome editing’ platforms enable robust activation or repression of transcription (CRISPR activation (CRISPRa) or CRISPR interference (CRISPRi), respectively) or tailored control over epigenetic modifications within human cells, which can be used to shape gene regulation and cell functions. d | The convergence of these transformative technologies can be used to engineer favourable properties within cell-based therapeutics, for example, by disrupting loci that elicit immunological recognition in therapeutic-grade induced pluripotent stem cells (iPSCs), overcoming limitations to therapeutic efficacy and natural potency by harmonizing integrated payloads with natural genetic regulatory programmes (that is, expressing a CAR-T receptor from a locus (TRAC) that naturally drives TCR expression) or overexpressing beneficial endogenous molecules, and by remodelling chromatin signatures to more efficiently reprogramme cellular lineage commitment, for instance, improving the derivation of iPSCs from fibroblasts. gRNA, guide RNA; PAM, protospacer adjacent motif; Transcript., transcriptional.