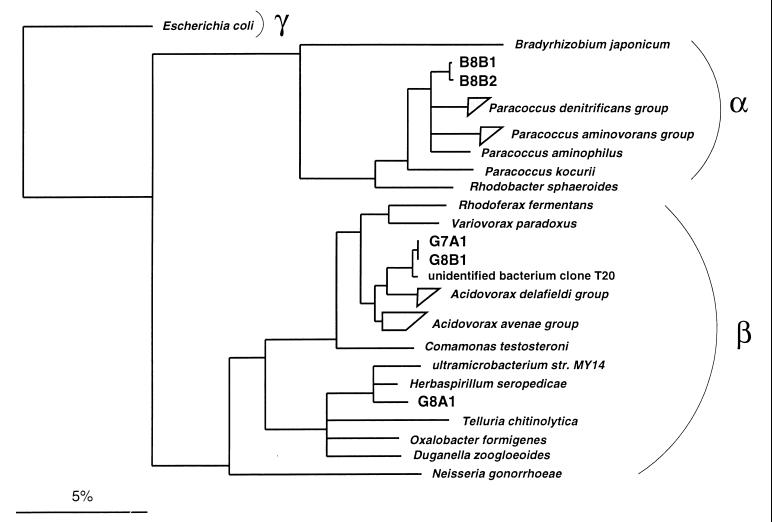
FIG. 3.
16S rRNA-based tree reflecting the phylogenetic relationships of (i) strains B8B1 and B8B2 and a selection of Proteobacteria from the alpha subclass and (ii) strains G7A1, G8B1, and G8A1 and a selection of Proteobacteria of the beta subclass. The tree is based on the results of a distance matrix analysis including complete or almost-complete 16S rRNA sequences from representative bacteria of the α, β, and γ subclasses (22). The topology of the tree was evaluated and corrected according to the results of distance matrix, maximum-parsimony, and maximum-likelihood analyses of various data sets. The phylogenetic positions of the analyzed strains did not differ in any of the treeing approaches. Multifurcations indicate topologies that could not be unambiguously resolved. The bar indicates 5% estimated sequence divergence. The P. denitrificans group comprises the species P. denitrificans (GenBank accession no. X69159) and P. versutus (D32243 and D32244) and Paracoccus sp. strains KL1, KS1, and KS2 (U58017, U58016, and U58015). The P. aminovorans group comprises the species P. aminovorans (D32240), P. alcaliphilus (D32238), and P. thiocyanatus (D32242). The A. delafieldii group comprises the species A. delafieldii (AF078764), A temperans (AF078766), and A. facilis (AF078765) and Acidovorax sp. strain 7078 (AF078767). The A. avenae group comprises A. anthurii (AJ007013), A. konjaci (AF078760), A. avenae subsp. avena (AF078759), A. avenae subsp. citrulli (AF078761), A. avenae subsp. cattleyae (AF078762), and Acidovorax sp. strain IMI 357678 (AF078763). Accession numbers of other bacterial 16S rRNA gene sequences are U00006 (Escherichia coli), D13429 (Bradyrhizobium japonicum), D32239 (P. aminophilus), D32241 (P. kocurii), X53855 (Rhodobacter sphaeroides), D16211 (Rhodoferax fermentans), D30793 (Variovorax paradoxus), Z93964 (unidentified bacterium clone T20), M11224 (Comamonas testosteroni), AB008503 (ultramicrobacterium strain MY14), Y10146 (H. seropedicae), X65590 (Telluria chitinolytica), U49757 (Oxalobacter formigenes), X74914 (Duganella zoogloeoides), and X07714 (Neisseria gonorrhoeae).