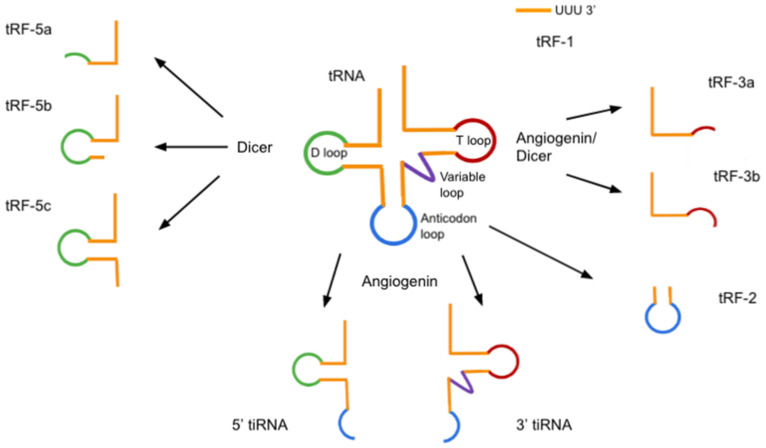
Figure 5.
Classification of tRFs and tiRNAs. tRF-1, also called 3′ U-tRF, is generated from the 3′ untranslated region during pre-tRNA maturation and is cleaved by RNase Z or its cytoplasmic homolog ribonuclease Z 2 (ELAC2) at the 5′ end. Therefore, the 3′ end of tRF-1 contains a poly U sequence. tRF-2 contains the anticodon loop and excludes the 5′ and 3′ end structures. tRF-2s are derived from tRNA-Glu, tRNA-Asp, tRNA-Gly, and tRNA-Tyr and are induced in hypoxic conditions. tRF-3s are generated from the 3′ end of mature tRNAs by cleavage in the T-loop by angiogenin or other members of the Ribonuclease A superfamily. tRF-3s can be further divided into two subclasses: tRF3a and tRF-3b, based on their lengths of either ~18 or ~22 nucleotides, respectively. tRF-5s are produced by Dicer cutting the D-loop or the stem position between the D-loop and the anticodon loop of the mature tRNA transcript. Depending on the cleavage sites and therefore different lengths, tRF-5s are further divided into three subtypes: tRF-5a (14–16 nt), tRF-5b (22–24 nt), and tRF-5c (28–30 nt). The oligonucleotide fragments shown as stem loops, for example, tRF-2, may or may not be thermodynamically stable but are drawn as such, primarily to show the tRNA regions they are derived from [166,168,169].