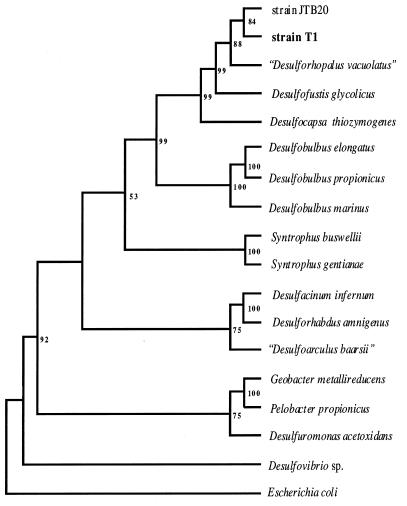
FIG. 3.
Neighbor-joining cladogram based on δ-proteobacterial 16S rDNA sequences. Sequences were aligned and edited in ClustalW. Hypervariable sites (as determined by less than 50% consensus of one base per alignment site) were removed. The aligned sequences were analyzed in PHYLIP. Seqboot was used to generate 500 bootstrap data sets. The bootstrap data sets were used to produce 500 distance matrices in DNADIST. The Kimura two-parameter model was used in DNADIST to account for probable variance in base substitution rates. Transversions were weighted twice as heavily as transitions were weighted. Neighbor-joining trees were constructed from the matrices by using NEIGHBOR. A consensus tree was generated in CONSENSE. The consensus tree was rooted with E. coli and was viewed in TREEVIEW. The numbers are bootstrap percentages. The levels of similarity between strain T1 and JTB20, “Desulforhopalus vacuolatus”, and Desulfofustis glycolicus were 95.3%, 93.0, and 91.5%, respectively.