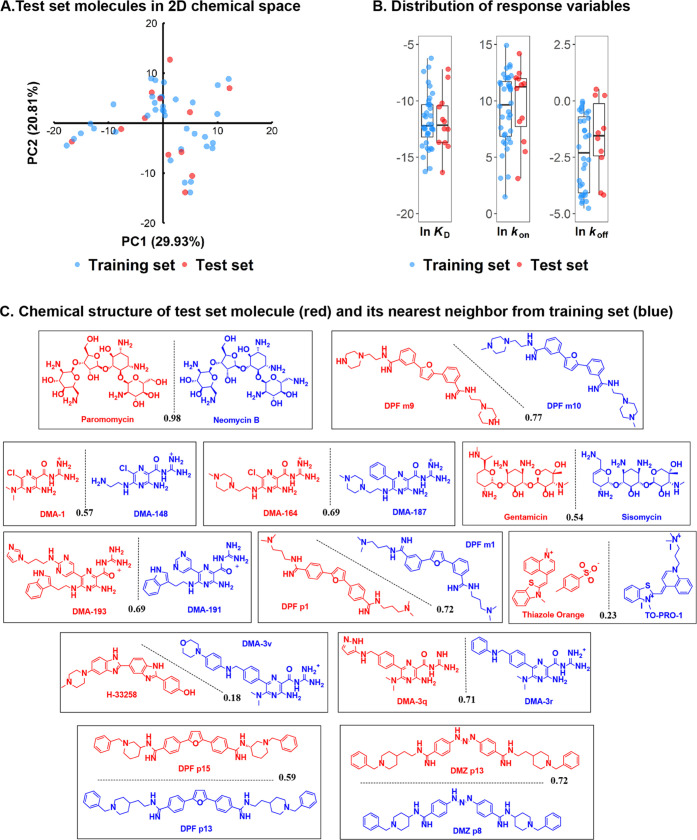
Figure 2.
A. Locations of test set molecules in the two-dimensional (2D) chemical space constructed from the first two principal components (29.9 and 20.8% of variances, respectively) of the whole data set. B. Distribution of response variables for the test and training set molecules. C. Chemical structures of the test set molecules (red) selected with the Kennard–Stone algorithm. The closest neighbor molecule in the training set (blue) is shown in pairs for comparison. The similarity was calculated as the Tanimoto coefficient (black) and is listed along the separation line.