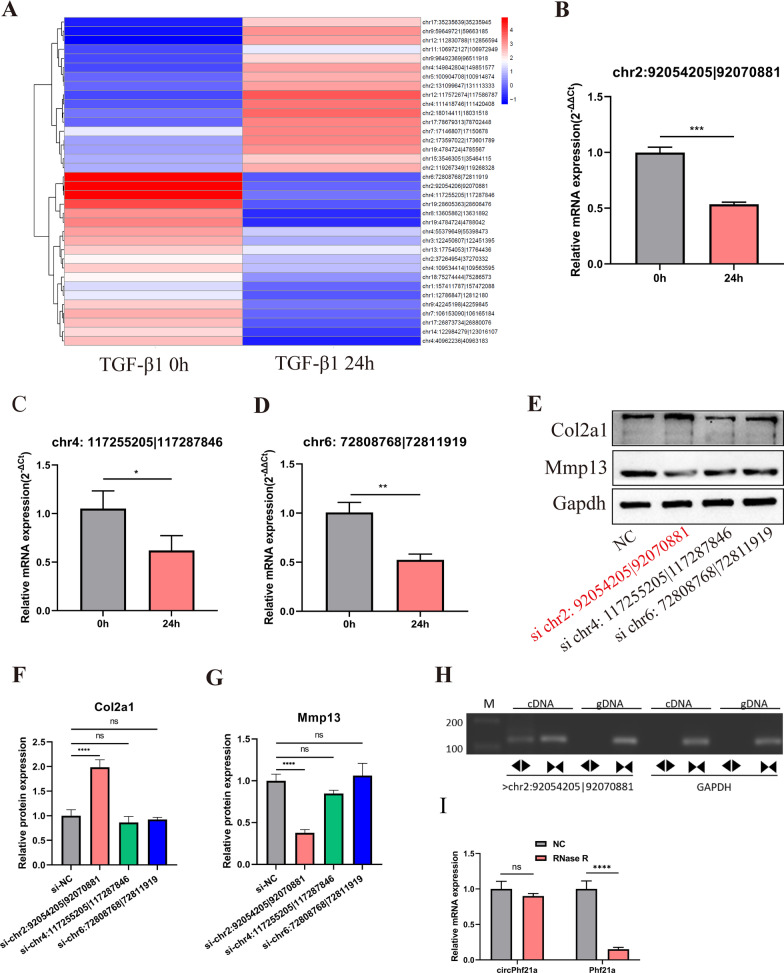
Fig. 2.
TGF-β1 related circRNA was identified by RNA-seq in PMCs. A Heatmap of all differentially expressed circRNAs, with | log2 (fold change) | > 1 and p < 0.05, between PMCs treated with 0 and 1 ng/ml TGF-β1 for 24 h. B–D Three most differentially expressed circRNAs were validated by RT-qPCR (n = 3, the results were consistent with those of sequencing), *p < 0.05, **p < 0.01, ***p < 0.001. E–G The protein expression levels of Col2a1 and Mmp13 were detected by western blotting after knockdown of chr2: 92054205|92070881 (circPhf21a), chr4: 117255205|117287846 and chr6: 72808768|72811919 in PMCs. (n = 3) ns p > 0.05, ***p < 0.001. H The presence of circPhf21a was validated in PMCs by RT-PCR. Divergent primers amplified circPhf21a from cDNA, but not from genomic DNA. Gapdh was used as a negative control. M = Marker. Back-to-back triangles represent divergent primers. Point-to-point triangles represent convergent primers. I The expression of circPhf21a and linear Phf21a mRNA in PMCs treated with or without RNase R was detected by RT-qPCR. The relative levels of circPhf21a and Phf21a mRNA were normalized to the values obtained with the mock treatment. n = 3 (three different experiments), ns p > 0.05, ****p < 0.0001