Fig. 4 |. Agonist discovery and the cryoEM structure of MRGPRX4.
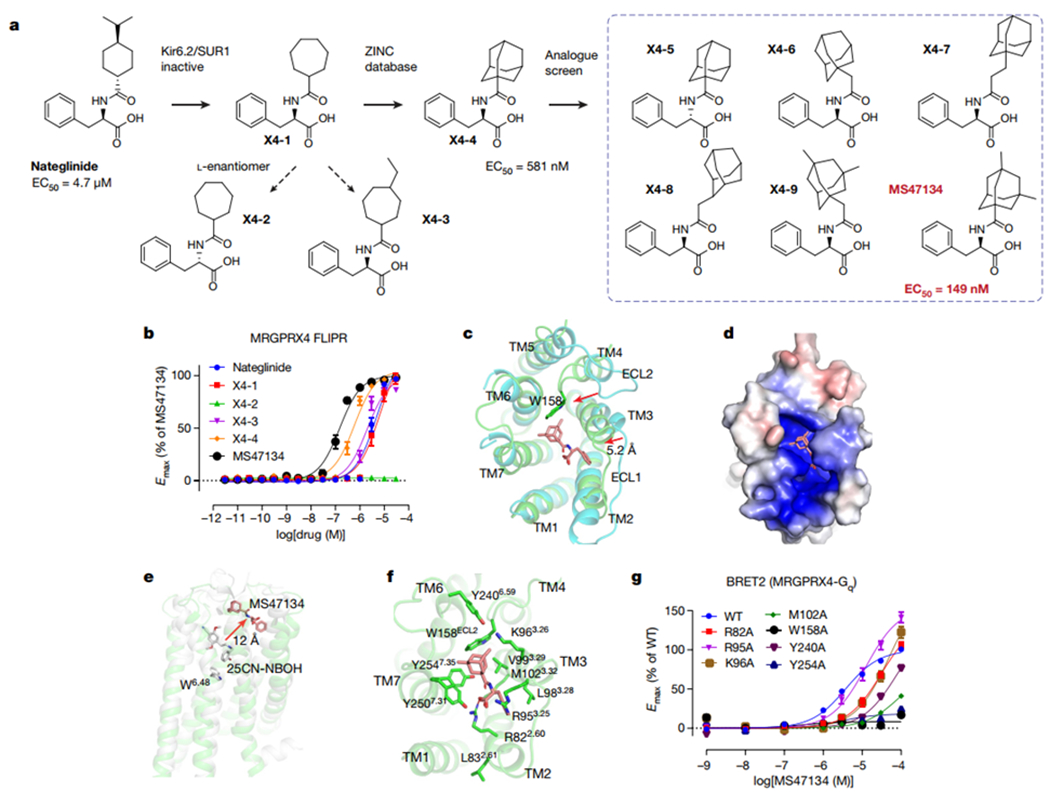
a, Overview of the compound optimization that leads to the discovery of MS47134. b, MS47134 displayed a significantly improved potency for MRGPRX4 in FLIPR Ca2+ assay compared with nateglinide. Data represent mean ± SEM of n = 3 biological replicates. c, Structural comparation of MRGPRX4 with MRGPRX2. Displacements of the extracellular part of MRGPRX4 related to MRGPRX2 are highlighted by red arrows. MRGPRX4 and MRGPRX2 are shown as green and cyan, respectively. MS47134 is shown as salmon sticks. d, Electrostatic surface representation of the MRGPRX4 extracellular pocket calculated using the APBS plugin in PyMOL. Red, negative (−5 kT/e); blue, positive (+5 kT/e). e, MS47134 binds to MRGPRX4 at the very extracellular side that is far away from the canonical toggle switch W6.48 in 5-HT2AR. MS47134, and 5-HT2AR agonist 25CN-NBOH are shown as sticks. f, Molecular interactions in the MS47134 pocket with surrounding residues shown as sticks. g, Alanine substitution on key residues in MS47134 pocket impaired MS47134 mediated Gq activation. Data represent mean ± SEM of n = 3 biological replicates.
