Extended Data Fig. 3 |. CryoEM images and data-processing of MRGPRX2-Gq-Cortistatin-14 and MRGPRX2-Gi1-Cortistatin-14 complex.
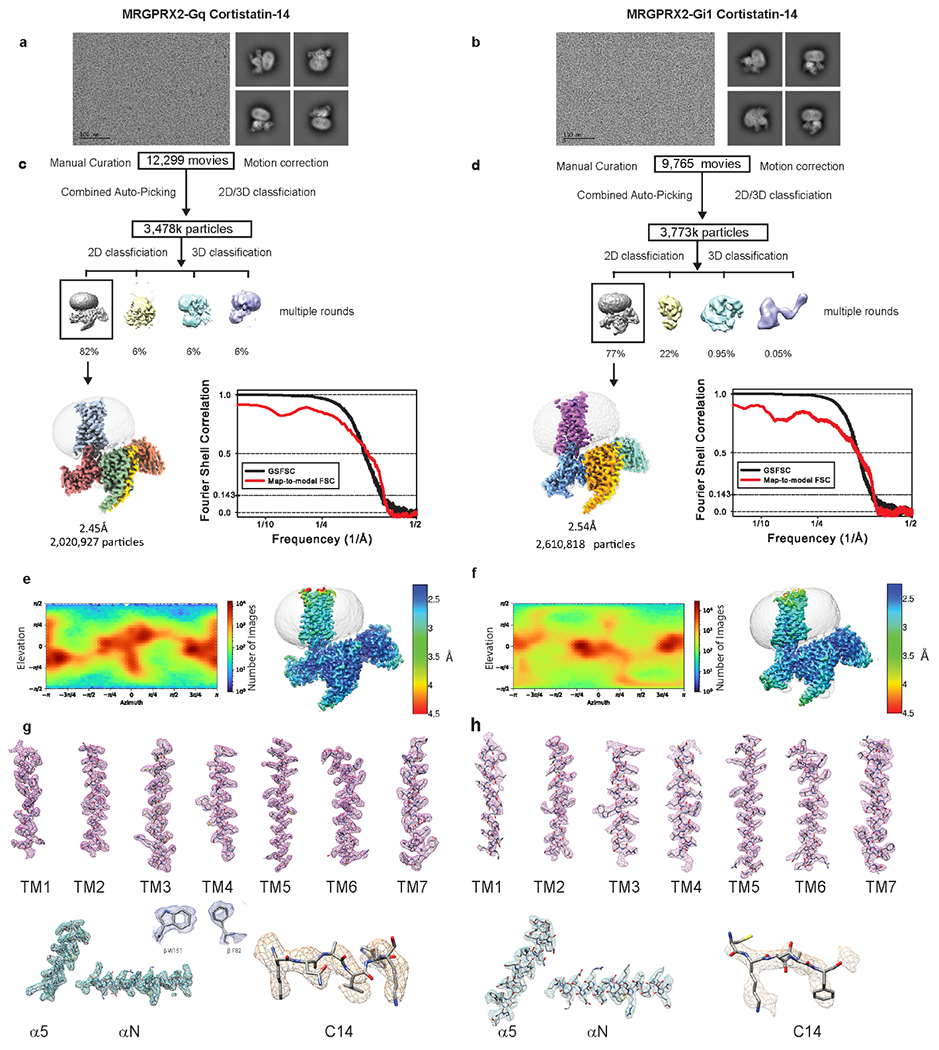
a-b, Representative motion corrected cryo-EM micrograph (scale bar, 100 nm) of MRGPRX2 G-protein cortistatin-14 (C14) particles imaged at a nominal 45k x magnification and representative two-dimensional class averages. The experiment was repeated three times with similar result. The exact number of movies and particles used for each complex are shown in the flow chart. c-d, Flow chart of cryo-EM data processing. GSFSC plot of auto-masked final map (black) and map-to-model real-space cross correlation (red) as calculated form phenix.mtriage. e-f, Viewing direction distribution and local resolution estimation heat maps. g-h, Local cryo-EM density maps of TM1-7, Cortistatin-14 ligand, α5 and α N helix of respective G-protein. Also shown inset are residues W151 and F82 of the b-subunit (blue).
