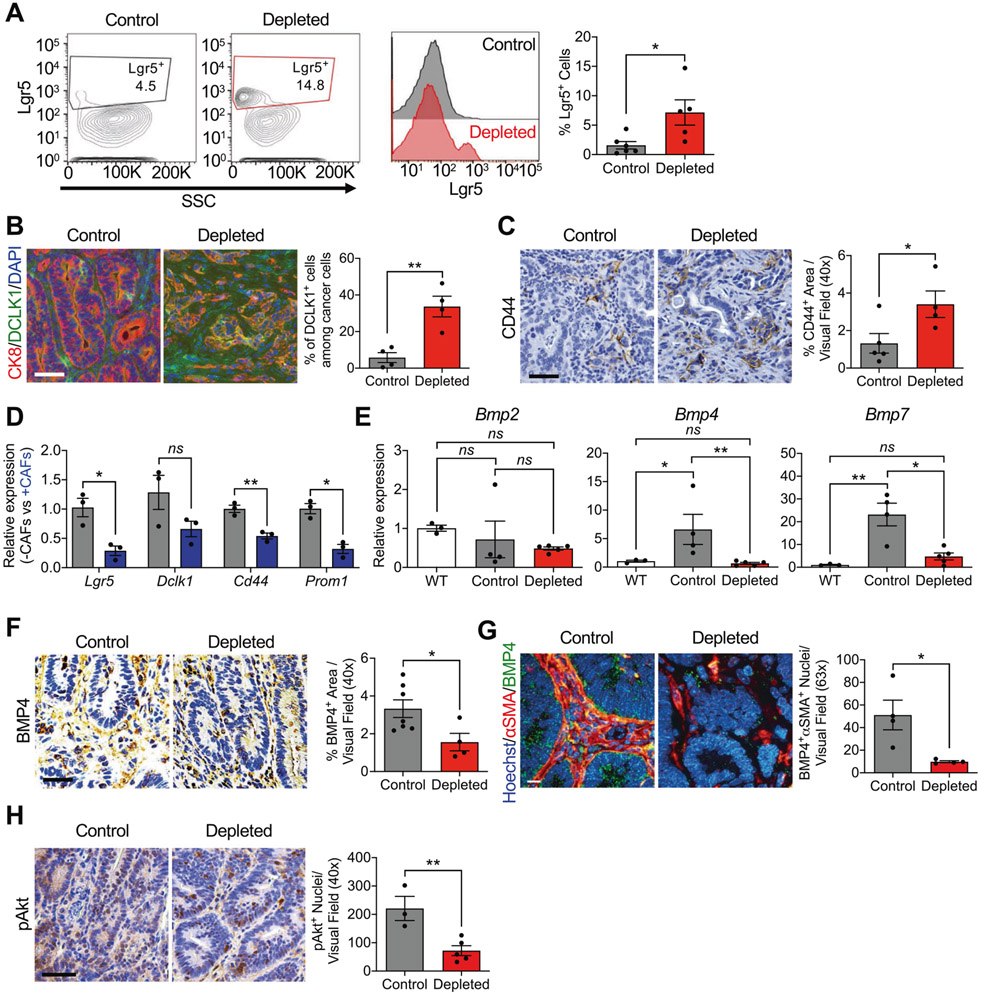
Fig. 3. αSMA+ CAF-depleted CRC tumors display an enhanced CSC phenotype.
A Representative contour plots, histograms, and quantification of Lgr5+ cells from control (black, n = 6) and depleted (red, n = 5) tumors. Number in gate indicate the % Lgr5+ cells. B Representative immunostaining of colon tumor sections for CK8 (red) and DCLK1 (green) in control (n = 4) and depleted (n = 4) tumors, and quantification of staining. DAPI (blue) was used as nuclear stain. Scale bar: 50 μm. C Representative immunohistochemistry of colon tumor sections for CD44 in control (n = 5) and depleted (n = 4) tumors, and quantification of staining. Scale bar: 50 μm. D Transcript levels of CSC markers Lgr5, Dclk1, Cd44, and Prom1 in cancer cells following co-culture with iKAP CAFs. n = 3 biological replicates. Statistics were performed based on ΔCT values. E Transcript levels of Bmp2, Bmp4, and Bmp7 in WT colon (n = 3), control tumors (n = 4), and depleted tumors (n = 5). F Representative immunohistochemistry of colon tumor sections for BMP4 and quantification of staining in control (n = 7) and depleted (n = 4) tumors. Scale bar: 50 μm. G Representative immunohistochemistry of colon tumor sections for BMP4 and αSMA, and quantification of staining of control (n = 4) and depleted (n = 4) tumors. Scale bar: 10 μm. H Representative immunohistochemistry of colon tumor sections for phosphorylated AKT (pAKT) and quantification of staining of control (n = 3) and depleted (n = 5) tumors. Scale bar: 50 μm. SSC: side scatter. The data are presented as mean ± SEM. A Mann–Whitney test performed; B, C, F–H unpaired t-test performed; D unpaired t-test with Welch’s correction performed; E one-way ANOVA with Sidak’s multiple comparison test performed based on ΔCT values. *P < 0.05, **P < 0.01, ns: not significant.