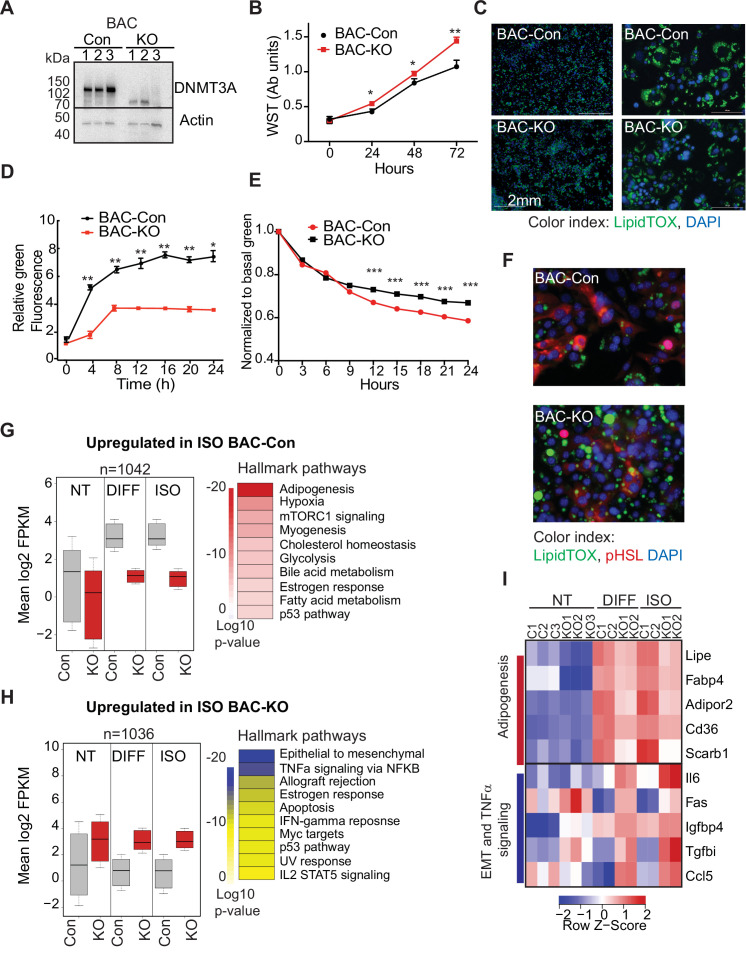
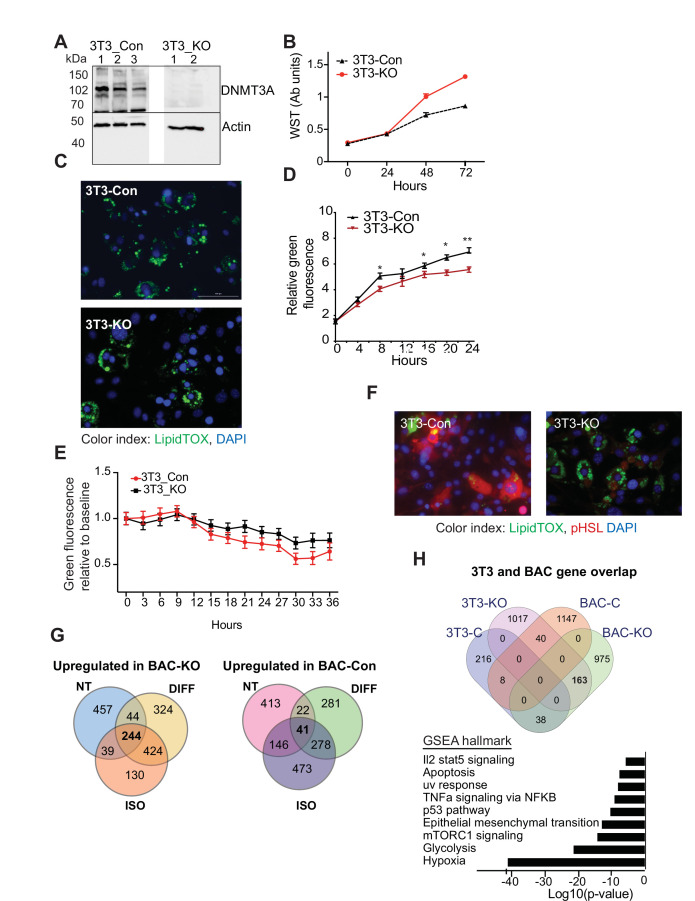
Figure 5. DNMT3A regulates preadipocyte proliferation and differentiation.
(A) Western blot probed with DNMT3A antibody after CRISPR KO in BAC-C4. Three clones of each are shown. (B) Proliferation of three WT and KO clones of BAC-C4 cells; assay based on water-soluble tetrazolium (MTT). (C) Representative staining of lipid droplets (LipidTOX (green)) following 6 days of differentiation of BAC-C4 control and KO cells. Scale 2 mm. (D) Incorporation of fluorescent fatty acid (BODIPY FL C12 (green)) relative to red nuclear fluorescent staining quantified during live cell imaging at day 5 of differentiation. Statistical analysis two-way ANOVA. *** p<0.0001 (E) Quantification of green, fluorescent fatty acid in control and KO cells following treatment with isoproterenol. Data shown is relative to the abundance of green fluorescence at baseline. Statistical analysis two-way ANOVA. *** p<0.0001 (F) Representative immunostaining of cells with the indicated antibodies. Cells were differentiated for 5 days and then treated with isoproterenol for 24 hr (G) RNAseq analysis at baseline (average of three clones from each genotype, non-treated, NT), at day 6 of differentiation (DIFF, two clones each genotype), and following 24 hr treatment of cells after 5 days of differentiation with isoproterenol (ISO, two clones each). Left panel: For genes defined as upregulated in BAC-control compared to KO following ISO, we plotted log transformed mean expression for all conditions. Right panel: Heatmap of MsigDB enrichment of genes upregulated in BAC-control (fold >2, p ≤ 0.05) compared to BAC-KO (Fold >2, p ≤ 0.005). (H) Using RNAseq data from (E): Left panel: log transformed mean expression for all conditions for genes defined as upregulated in BAC- KO compared to control following ISO. Right panel: Heatmap of MsigDB enrichment of genes upregulated in BAC-KO (fold >2, p ≤ 0.05) compared to BAC-Control (Fold >2, p ≤ 0.005). (I) Heatmap of top 5 genes differentially expressed between control and KO and annotated to the hallmarks adipogenesis or Epithelial to mesenchymal transition (EMT) and TNFa signaling pathways.