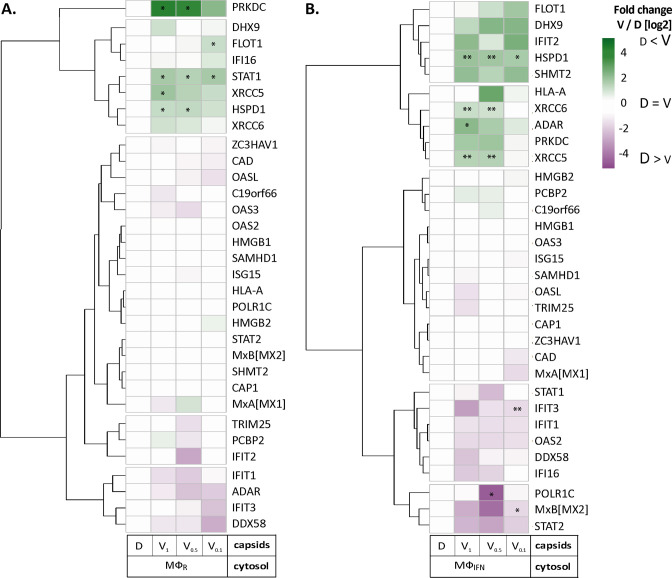
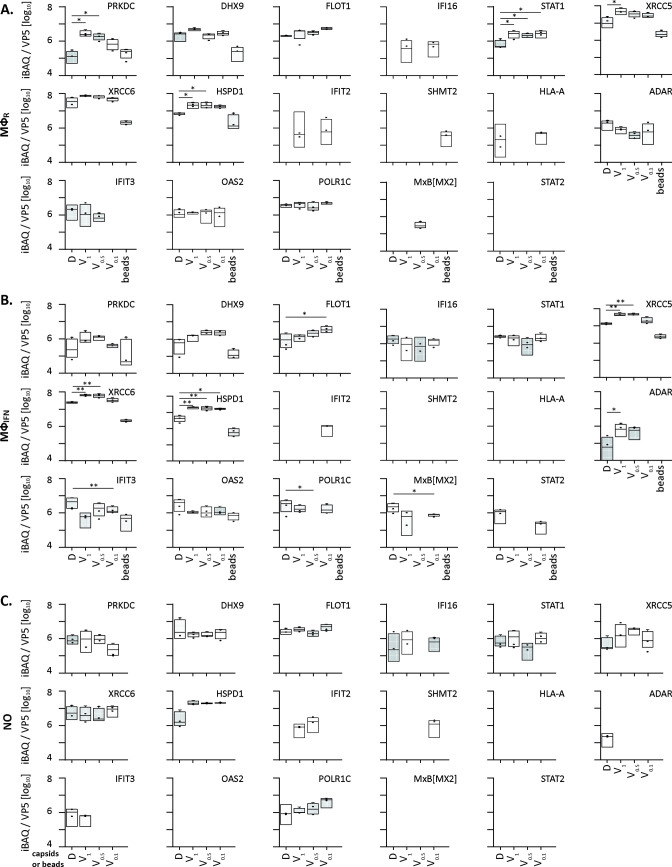
Figure 4. HSV-1 capsids associate with proteins involved in type I IFN response.
Unbiased hierarchical clustered heat map showing the log2 fold changes of IFN-induced proteins (GO type-I IFN) identified from capsids-host protein sediments from cytosol of resting Mφ, or IFN-induced MφIFN macrophages. For each protein, the fold change was calculated based on their abundance (iBAQs) in V1, V0.5, and V0.1 capsids as compared to their abundance in D capsids, using a linear scale from violet being the lowest to dark green being the highest. (*) and (**) design the proteins with an FDR corrected p-value ≤ 0.05 and ≤ 0.01, respectively.