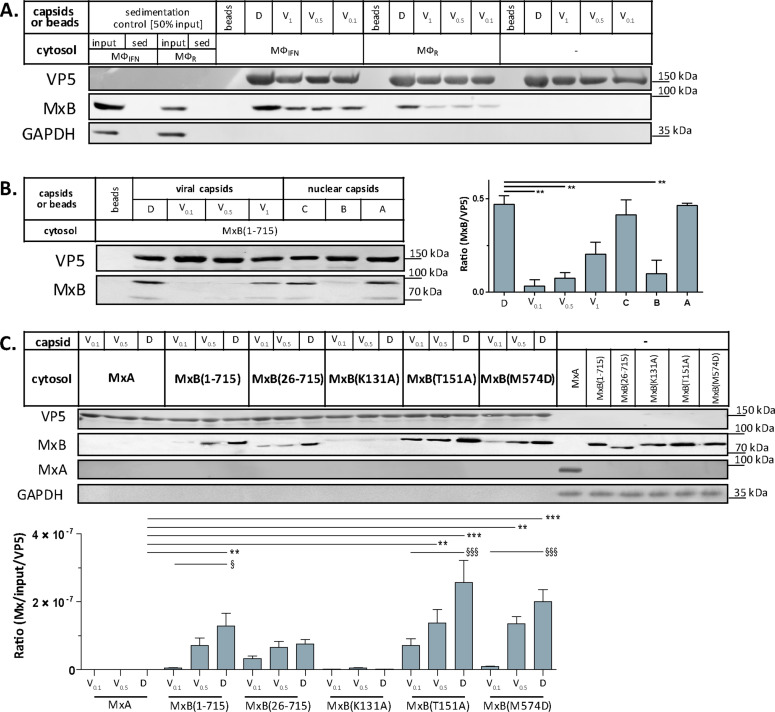
Figure 5. Tegumentation reduces MxB binding to HSV-1 capsids.
The binding of MxB to viral V0.1, V0.5, V1, or D, or to nuclear A, B, or C capsids was analyzed after incubation in 0.2 mg/mL cytosol prepared from (A; Figure 5—source data 1) THP-1 φ stimulated or not with IFN, or (B-C; Figure 5—source data 1; Figure 5—source data 1) A549 cells stably expressing MxA, MxB(1-715) full length, the short MxB(26-715), or MxB mutants defective in GTP-hydrolysis MxB(T151A), GTP-binding and hydrolysis MxB(K131A), or dimerization MxB(M574D). Sedimented capsid-host protein complexes were then analyzed by immunoblot for VP5 (capsid), MxB, MxA, and GAPDH as a loading control. As control cytosols were sedimented without capsids (A: sed), or with uncoated agarose beads (A, B: beads). The amounts of MxA/MxB found in the capsid-host protein complexes were quantified, and normalized to their respective VP5 levels. Error bars: SEM. summarized from three experiments. One asterisk denotes p < 0.05, two asterisks indicate p < 0.01 and three asterisks represent p < 0.001 as determined by Welch’s t-tests comparisons.