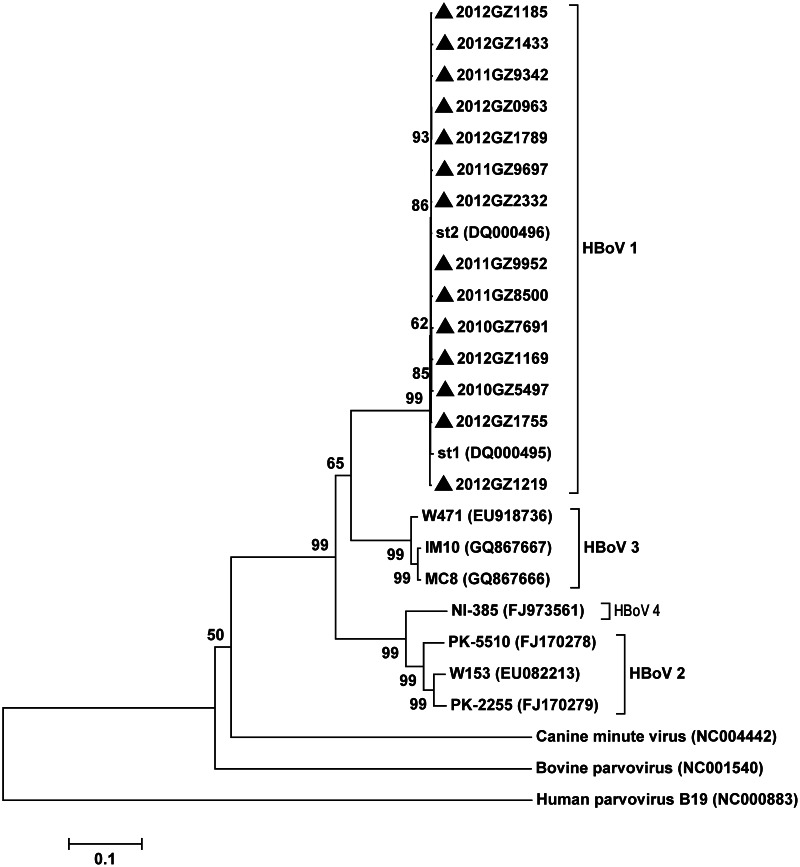
Fig. 3.
Phylogenetic tree constructed from the complete genomes of 14 human bocavirus strains identified in faecal samples, along with other representative bocavirus genotypes available in the GenBank database. Phylogenetic trees were inferred from complete genome data with 1000 bootstrap replicates using the neighbour-joining method with Mega 5·2 software. The scale bar indicates the estimated number of substitutions per 500 bases by Kimura's two-parameter model. Parvovirus B19, bovine parvovirus, canine minute virus, and representative strains of HBoV1–4 were used as reference strains for genotype analysis of 14 human bocavirus strains identified in faecal samples (labelled with a black triangle). The 14 HBoV strains obtained in this study were identified as HBoV1.