SUMMARY
The faecal-pat prevalence (as estimated by culture) of Campylobacter fetus from cattle and sheep on 19 farms in rural Lancashire was investigated using standard Campylobacter culture techniques and PCR during a 2-year longitudinal study. C. fetus was isolated from 9·48% [95% confidence interval (CI) 8·48–10·48] of cattle faecal pats and 7·29% (95% CI 6·21–9·62) of sheep faecal pats. There was evidence of significant differences in shedding prevalence between geographical regions; cows in geographical zone 3 had an increased risk of shedding C. fetus compared to cows in geographical zones 1 and 2 (OR 6·64, 95% CI 1·67–26·5, P = 0·007), as did cows at pasture (OR 1·66, 95% CI 1·01–2·73, P = 0·046) compared to when housed. Multiple logistic regression modelling demonstrated underlying seasonal periodicity in both species.
Key words: Campylobacter, veterinary epidemiology, zoonotic foodborne diseases
INTRODUCTION
Bacteria belonging to the genera Campylobacter are resident intestinal flora of many birds and mammals, with some species such as Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus recognized globally as significant human and animal pathogens [1]. Of particular concern, is enteritis caused by Campylobacter spp. which is a major cause of foodborne enteritis worldwide and in the UK [2]. The epidemiology of Campylobacter infection is complex and a number of animal species, especially food animals such as poultry and ruminants, are implicated as sources of human disease through foodborne and environmental transmission pathways [3].
In comparison with other Campylobacter spp., C. fetus is a less well studied cause of human zoonotic disease. It has two subspecies, C. fetus subsp. fetus and C. fetus subsp. venerealis [4]. Infection in humans is usually but not exclusively [5] associated with C. fetus subsp. fetus which causes a range of disease including bacteraemia, enteritis, vascular disease, endocarditis, abortion, infertility and meningo-encephalitis, especially in the immunocompromised, the elderly and the young [3]. It has been proposed that the source of human infection with C. fetus is zoonotic, through contamination of food [3]. Human infections have been linked to consumption of raw meat [6, 7], raw milk [8] and working in abattoirs [9]. However, this does not explain C. fetus infections in all cases [10]. Furthermore, the ability C. fetus to survive for up to several weeks in a range of biological materials, including faeces and water, demonstrates the potential for contaminated environments to act as a source of human infection [11]. However, further epidemiological investigations into environmental and foodborne routes of human infection are required.
As well as being a human pathogen, C. fetus subsp. fetus has also been long recognized as an important and common cause of abortion and fetal infection in sheep [12]. For example, in the UK in 2011, Campylobacter spp., chiefly C. fetus, were isolated from 6·6% of reported abortions in sheep [13]. Infection of sheep is thought to occur by ingestion of food or water contaminated by sheep or wild-life faeces [14]. C. fetus subsp. venerealis is a common cause of abortion and infertility in cattle worldwide. It is adapted to the bovine genital tract, venerally transmitted and has been reported as a cause of human disease [5, 15].
There are few data on the epidemiology of C. fetus in farm animals. Such information is required for the full elucidation of the role of farm animals in the epidemiology of human infection. The aim of the current study was to investigate the prevalence, and seasonal and farm management factors associated with C. fetus shedding in cattle and sheep in a cohort of dairy and sheep farms in the Northwest of England. The current study is nested in a larger study of the molecular epidemiology of C. jejuni in this cohort of farms [16, 17].
MATERIALS AND METHODS
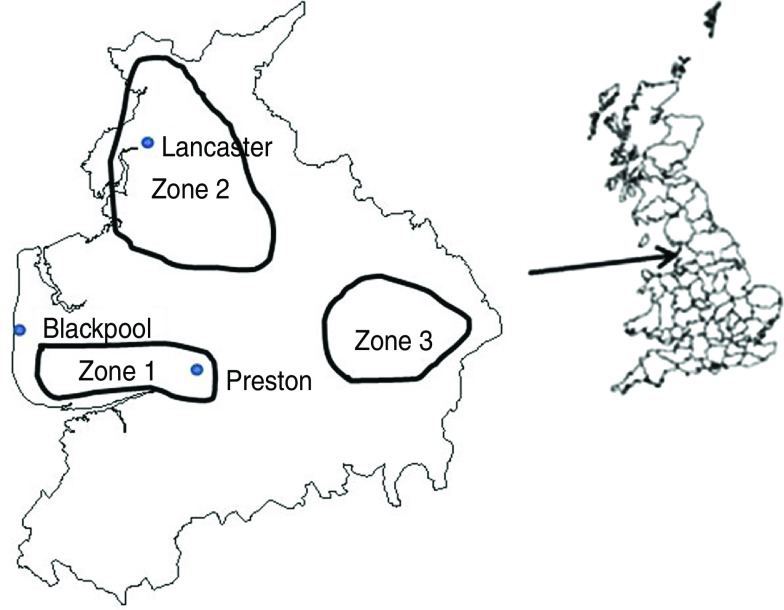
The study design was a repeated cross-sectional study of 15 dairy and four sheep farms in Lancashire. Farms were recruited via three geographically distinct veterinary practices in Lancashire serving Fylde (zone 1), North Lancashire (zone 2) and South East Lancashire (zone 3) (Fig. 1). Farms were visited at 8-week intervals when 20 freshly voided faecal samples were collected from adult animals. In the case of bovine faeces consistency was scored [18], and a larger sample was collected from the same faecal pat for further analysis by sieving to assess fibre length and presence of partially digested grains [19].
Fig. 1.
Map of Lancashire, UK, showing geographical location of sampling sites.
Since the primary aim of the study was the investigation of the epidemiology of C. jejuni, culture methods were optimized for isolation of C. jejuni. One gram of faeces was inoculated into 9 ml Campylobacter enrichment broth (IDG Ltd, UK) with cefoperazone, vancomycin, trimethoprim and cyclohexamide (CVTC supplement, IDG Ltd) and incubated for 24 h at 37 °C in a microaerobic atmosphere (12% CO2, 3% H2, 11% O2, 74% N2). After incubation, two aliquots (50 μl and 5μl) of the enrichment broth were inoculated onto two Campylobacter blood-free selective agar (CSA) plates (IDG Ltd) enriched with cefoperazone and amphotericin (CA supplement, IDG Ltd, and incubated at 37°C in a microaerobic atmosphere for 60–72 h. Up to four putative Campylobacter colonies (per faecal sample) were serially subcultured twice onto blood agar plates and incubated at 37°C for 72 h each time under microaerobic conditions. Following culture, a crude DNA aqueous lysate was prepared from putative Campylobacter isolates by inoculating 200 μl distilled water with a small amount of the culture, heating at 100°C for 15 min followed by centrifugation at 1137 g for 10 min. Assignment to species of putative Campylobacter spp. was by PCR using the following assays: a 16S rRNA PCR for identification of Arcobacter spp. [20]; a multiplex PCR for identification of C. jejuni, C. coli and C. lari [21]; a monoplex PCR [22] for identification of any C. jejuni that failed to be identified by the multiplex PCR and a 16S rRNA-based duplex PCR for identification of C. fetus and C. hyointestinalis [23].
A random sample of 30 bovine isolates was selected for multilocus sequence typing (MLST) as described previously [24]. Sequencing was performed using an ABI 3130xl automatic sequencer with a 50-cm array and POP-7 polymer (Applied Biosystems, USA). Sequence data assembly, alignment, and interrogation of the C. fetus MLST website (http://pubmlst.org/campylobacter/) for assignment of sequence type was performed using the STARS computer program [25].
Data analysis was performed using Stata v. 12 (StataCorp., USA). Covariates recorded at sampling visits and considered for inclusion in statistical analyses are described in Table 1. The sampling period was split into ‘summer’ and ‘winter’ with the winter period defined as being from 1 October to 30 April. The term ‘sampling event’ is defined as ‘a visit to a farm to collect samples’. The continuous variables, of milk yield and group size were transformed into quintiles with stocking density (sheep) being transformed into quartiles.
Table 1.
Description of variables collected at sampling visits for initial inclusion in statistical analyses
| Variables | Species | Type | Description and coding of variable |
|---|---|---|---|
| Farm identity | Cattle/sheep | Categorical | |
| Purchase policy | Cattle | Categorical | 0 = No purchased stock (closed herd) |
| 1 = Occasional purchase of cows | |||
| 2 = Frequent purchase | |||
| Group size | Cattle/sheep | Continuous | Number of animals in sampled group (transformed into quintiles) |
| Where sampled | Cattle/sheep | Categorical | Inside = 1; outside = 0 |
| Zone | Cattle/sheep | Categorical | 1 = Southern Fylde |
| 2 = North Lancashire | |||
| 3 = South East Lancashire | |||
| Date of sampling | Cattle/sheep | Dd/mm/yy | |
| Average daily milk yield | Cattle | Continuous | Average daily milk yield (litres) on sampling day (transformed into quintiles) |
| Feeding system | Cattle | Categorical | Feeding system used as follows: |
| 1 = TMR – total mixed ration | |||
| 2 = Hybrid TMR – TMR and parlour feed | |||
| 3 = Grazing and buffer feed and parlour feed | |||
| 4 = Grazing and parlour feed | |||
| 5 = Silage and parlour feed | |||
| Number of fresh cows | Cattle | Continuous | Number of cows calved within last month (transformed into quintiles) |
| Faecal score | Cattle | Categorical Binary | Score of 1–5 depending on consistency with score 1 being very firm and score 5 being liquid [17]; firm = score 2 or 3, loose = score 4 or 5 |
| Sieve score | Cattle | Binary | Score being a composite score for presence of grains and long fibres with 0 = no (or a few) grains or long fibres; 1 = large amounts of grains and presence of many long fibres [18] |
| Stocking density | Sheep | Continuous | Number of sheep per hectare (transformed into quartiles) |
| Pasture quality | Sheep | Categorical | Quality of pasture, scored 0 or 1 (0 = poor; 1 = mediocre to lush) |
| Lambing season | Sheep | Categorical | Were the flock lambing at the time of sampling? |
| 1 = sampled in lambing season | |||
| 0 = sampled out of lambing season |
The technique of selecting four isolates per plate and testing each independently for a number of species of the family Campylobacteraceae introduces a dependency in the sensitivity of isolation for each species, in that the probability of isolating a specific organism, e.g. C. fetus from a faecal pat co-colonized by another organism, e.g. C. jejuni will depend on the relative frequency of each organism within the pat. Thus isolation of a C. jejuni would reduce the probability of isolating a C. fetus and vice versa. To account for this dependency a binary variable ‘other campylobacter present’ was included as a covariate in the multivariable models.
Following univariate analysis, performed using χ2 test and logistic regression to investigate associations between C. fetus faecal-pat prevalence and the measured covariates, multiple logistic regression models were fitted for cattle and sheep with the binary outcome variable being the C. fetus test result (presence or absence) of the faecal-pat sample. Explanatory variables under consideration are given in Table 1 while time was forced into the models. Time was considered as a composite of four sine and cosine functions (harmonic regression) to allow modelling of seasonal periodicity if present. Four time covariates (x1, x2, x3, x4) were generated as follows:
where t = week number with week 1 being the first week in January 2006 when sampling commenced. The impact of season on the prevalence of Campylobacter in discrete faecal pats, after adjusting for the other covariates in the model, was demonstrated visually by plotting the logit predictions using the time covariates only.
A backward elimination strategy was employed whereby a full model was built and then each variable removed in turn, a likelihood ratio test performed and the resultant P value noted. The variable with the highest P value was then omitted and the process repeated. This process was repeated until only variables with P < 0·05 remained in the model. The omitted variables were then added back in turn, starting with the lowest P value, a likelihood ratio test performed after each addition, and the variable retained if P < 0·05. This process was continued until no further variables could be added, to produce the final model. Interactions between variables in the final model were considered for inclusion and retained if they improved model fit as judged by the likelihood ratio test. No significant interactions were identified.
To account for the hierarchical nature of the dataset, a random-effect logistic regression model was fitted with farm considered as a random effect in the case of the cattle dataset. A logistic regression model with farm identity as a fixed effect was fitted for the sheep dataset since there were only four sheep farms sampled. In both cases, the underlying a priori hypothesis was that time of year or season is a primary determinant of the probability of a faecal pat being colonized by C. fetus with other covariates also having an effect. Estimates of C. fetus faecal-pat prevalence were obtained from regression models.
RESULTS
Study farm details
The median herd size, defined as total number of lactating and dry cows, was 145 [interquartile range (IQR) 104–200]. Four herds were housed all the time during the study period while one was housed for the entire second year of the study. All other herds were housed during the winter months but grazed outside during the summer. Median annual milk yield was 8000 (IQR 7200–9000) litres per cow. Two of the sheep farms were lowland, with one farm grazing on the salt-marshes of a river estuary, while two were upland with one utilizing summer grazing on moorland. The predominant breeds of sheep kept were Swaledale and North Country Mules. The two upland farms kept 1000 and 700 ewes, respectively, while one lowland farm had 700 ewes and the other 150.
Campylobacter spp. isolation
Twenty faecal samples were collected at each sampling visit, yielding a total of 4260 samples. Four potential isolates were taken from each faecal sample yielding 17 040 potential bacterial isolates. In total, 9499 (55·75%) putative Campylobacter spp. were isolated from faecal pats from both species with 1082 (11·39%) isolates identified as C. fetus (Table 2).
Table 2.
Distribution of Campylobacter spp. by host species [15]
| Cattle | Sheep | |
|---|---|---|
| Number of faecal-pat samples | 3300 | 960 |
| Number of potential isolates (4 per pat) | 13 200 | 3840 |
| Number of isolates grown (% of potential isolates) | 7779 (58·9%) | 1720 (44·8%) |
| Campylobacter jejuni (% of actual isolates) | 1857 (23·9%) | 450 (26%) |
| Campylobacter coli (% of actual isolates) | 346 (4·4%) | 815 (47·4%) |
| Campylobacter fetus (% of actual isolates) | 871 (6·8%) | 211 (12·3%) |
| Campylobacter hyointestinalis (% of actual isolates) | 380 (4·9%) | 0 |
| Campylobacter lari (% of actual isolates) | 26 (0·33%) | 8 (0·05%) |
At farm level, C. fetus was present on all farms with the exception of one cattle and one sheep farm. C. fetus was isolated on 64% of cattle farm visits and 37·5% of sheep farm visits. C. fetus was isolated from 9·48% (95% CI 8·48–10·48) of cattle faecal pats and 7·92% (95% CI 6·21–9·62) (Table 3) of sheep faecal pats (Table 4). There was no species difference in prevalence (P = 0·138).
Table 3.
Random-effects multivariable logistic regression model including covariates associated with the probability of isolating Campylobacter fetus spp. from cattle faeces on Lancashire dairy farms. Farm is considered as a random effect (n = 15). Coefficients for time covariates are not included in Table 3
| Covariate | Estimate β | 95% CI | OR | 95% CI | Wald test P value |
|---|---|---|---|---|---|
| Pasture vs. housed | 0·51 | 0·01 to 1·00 | 1·66 | 1·01–2·73 | 0·046 |
| Geographical zone | |||||
| Zone 2 vs. zone 1 | 0·69 | −0·64 to 2·03 | 2·0 | 0·53–7·6 | 0·31 |
| Zone 3 vs. zone 1 | 1·89 | 0·51 to 3·28 | 6·64 | 1·67–26·5 | 0·007 |
| Presence of long fibres in faeces | 0·74 | 0·01 to 1·00 | 2·10 | 1·30–3·40 | 0·046 |
| Other bacteria isolated from same faecal pat | −0·97 | −1·23 to −0·71 | 0·38 | 0·29–0·49 | <0·0001 |
| Baseline: housed in zone 1 with no long fibres in faeces and no other bacteria isolated | −3·36 | −4·32 to 2·40 | |||
OR, Odds ratio; CI, confidence interval.
Between-farm variance 0·25 (95% CI 0·12–0·43).
Table 4.
Fixed-effects multivariable logistic regression model including covariates associated with the probability of isolating Campylobacter fetus spp. from sheep faeces on Lancashire sheep farms. Farm is considered as a fixed effect (n = 4). Coefficients for time covariates are not included in Table 4
| Covariate | Estimate β | 95% CI | OR | 95% CI | Wald test P value |
|---|---|---|---|---|---|
| Farm | |||||
| Farm 9 vs. zone 15 | 1·50 | 0·74 to 2·26 | 4·5 | 2·1–9·6 | <0·0001 |
| Farm 12 vs. zone 15 | 1·06 | 0·26 to 1·85 | 2·87 | 1·30–6·34 | <0·0001 |
| Other bacteria isolated from same faecal pat | −0·76 | −1·32 to −0·20 | 0·47 | 0·27–0·81 | 0·007 |
| Baseline: farm 15 and no other bacteria isolated | −2·91 | −3·64 to −2·19 | |||
OR, Odds ratio; CI, confidence interval.
MLST demonstrated little diversity with four sequence types being identified, namely ST2 (n = 5), ST3 (n = 19), ST5 (n = 5) and ST6 (n = 1). These all belong to subspecies C. fetus fetus.
Univariate analysis
For cattle, C. fetus faecal-pat prevalence, was higher in geographical region zone 3 (OR 1·65 95% CI 1·43–1·90, P < 0·001) compared to the other zones. Testing of cattle sampled at pasture, compared to housed cattle, revealed a higher faecal-pat prevalence at pasture (OR 1·53, 95% CI 1·21–1·94, P < 0·001). There was no difference in faecal-pat prevalence of C. fetus in summer compared to winter (P = 0·61). Both milk yield (quintiles) and group size (quintiles) were negatively associated with C. fetus pat prevalence (OR 0·61 95% CI 0·55–0·69, P < 0·001 and OR 0·78, 95% CI 0·70–0·89, P < 0·001 respectively). In sheep, the faecal-pat prevalence of C. fetus was higher in summer compared to winter (OR 2·77 95% CI 1·68–4·47, P < 0·001). No association between C. fetus pat prevalence and any other management factor was observed.
Multivariable analysis
Cattle: random-effects logistic regression model (Table 3)
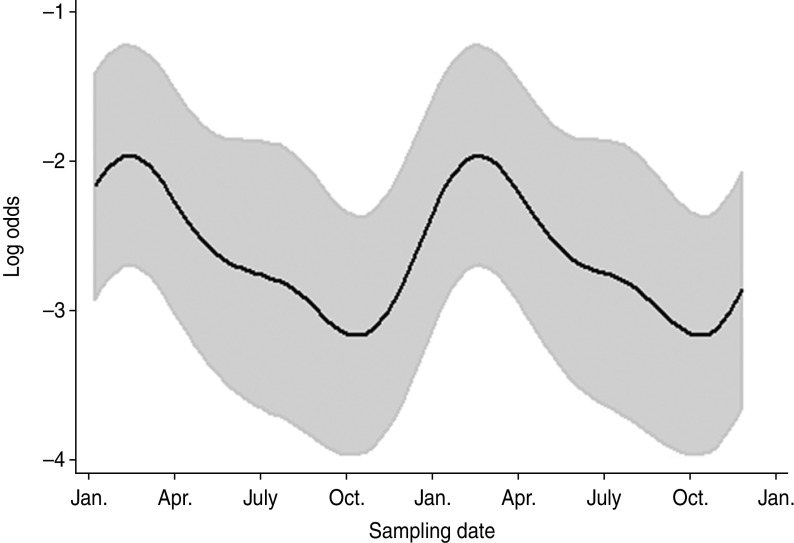
Farm identity was included as a random effect. A strong positive association between C. fetus faecal-pat prevalence and geographical zone 3 was observed (OR 6·64, 95% CI 1·67–26·5, P = 0·007). Cattle at pasture had an increased risk of shedding C. fetus compared to when they were housed (OR 1·66, 95% CI 1·01–2·73, P = 0·046). The presence of long fibres in the faeces (faecal sieve score) was associated with an increased probability of isolating C. fetus (OR 2·1, 95% CI 1·30–3·40, P = 0·007). The seasonal influence on faecal-pat prevalence, after adjusting for the other covariates in the model is shown in Figure 2, with a peak observed in February in both years.
Fig. 2.
The seasonal component to variation in Campylobacter fetus faecal-pat prevalence on Lancashire dairy farms (n = 15). The grey areas represent the upper and lower 95% confidence intervals.
The variance associated with farm identity in faecal-pat prevalence of C. fetus due to farm effect was estimated to be 0·25 (95% CI 0·12–0·43).
Sheep: fixed-effects logistic regression model (Table 4)
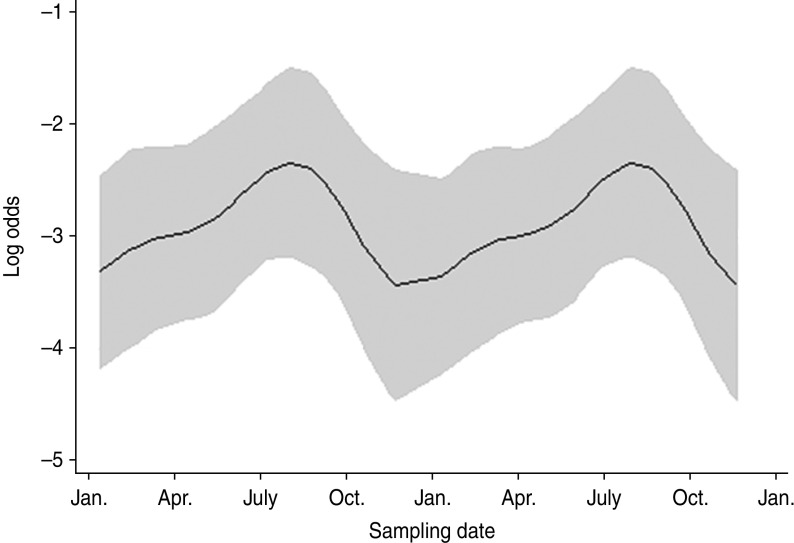
Farm identity was considered a fixed effect with farm no. 15 included as the baseline (as C. fetus was never isolated from farm no. 16. No associations were found between C. fetus faecal-pat prevalence and any recorded farm management practices. The sesonal influence on faecal-pat prevalence after adjusting for farm identity is shown in Figure 3, with peaks during early spring and late autumn.
Fig. 3.
The seasonal component to variation in Campylobacter fetus faecal-pat prevalence on Lancashire sheep farms (n = 4). The grey areas represent the upper and lower 95% confidence intervals.
DISCUSSION
In interpreting the results of the present study, it must be borne in mind that the microbiological techniques employed were optimized for the isolation of C. jejuni and not for C. fetus. It is well recognized that inclusion of antibiotics such as cefoperazone will inhibit growth of some C. fetus strains [26] allowing overgrowth by more robust species such as C. jejuni. Thus it is possible that the results presented are skewed towards detection of certain strains. Diversity in C. fetus as demonstrated by MLST is considerably less than that observed in C. jejuni, with 44 sequence types currently recorded on the ‘pubmlst database’ at the present time (http://pubmlst.org/campylobacter; accessed 14 May 2013). In the current study, only four sequence types (STs 2, 3, 5 6) were identified from 30 bovine isolates. Whether this limited range of sequence types isolated represents the true diversity present or is resultant on the use of suboptimal culture techniques cannot be determined. Furthermore, it must be realized that since only four isolates per faecal sample were speciated, the likelihood of detection of C. fetus will be influenced by the relative frequency of other Campylobacter spp. in the faecal sample. We accounted for this dependency in our multivariable analyses but could not account for the possibility of overgrowth by more robust species. Consequently, we suggest that the prevalence estimates presented are best considered as minimum prevalence estimates, derived using a test of imperfect sensitivity, i.e. the estimates presented are probably biased downwards.
This study demonstrates that cattle and sheep are significant reservoirs of C. fetus with all except one dairy farm and one sheep farm sampled showing evidence of colonization. All the sequence types isolated in our study have been associated with human disease with ST3 being the most frequently isolated sequence type from human cases in the UK [27]. Our findings would confirm the hypothesis that ruminants and their environment are potential sources of human infection with C. fetus.
In the case of dairy cattle, peak faecal-pat prevalence was observed during the spring while in sheep prevalence was highest during the summer months. Regression modelling suggested that in addition to an underlying seasonal pattern there was also an effect of sampling environment, with higher prevalences associated with cattle being at pasture compared to being housed. We observed a similar pattern in the epidemiology of C. jejuni on the same study farms [16]: increased faecal-pat prevalence associated with grazing pasture rather than season per se. In the present study four of the dairy farms housed lactating cattle throughout the year with no access to pasture. This practice is gaining in popularity in the UK due to the greater nutritional demands placed upon the high yielding Holstein dairy cow which are impossible to meet at pasture. The data presented here on C. fetus and previously on C. jejuni [16] faecal-pat prevalence may suggest that housing cattle would result in lower levels of excretion offering a means of reducing environmental load and associated human infection risk.
In temperate countries, the incidence of human campylobacteriosis is associated with temporal factors [28], with peak incidence in the UK occurring during the summer. The reasons for this seasonality remain unknown. Seasonal trends have also been observed in poultry and ruminant Campylobacter shedding [29], with cattle showing two peaks in spring and autumn, and a spring peak in slaughter lambs [30]. More specifically, a strong seasonality has been observed for C. jejuni shedding from dairy cattle [16]. A number of reasons for these seasonal trends have been proposed, including climate [31], and farm management factors such as nutrition, housing, grazing patterns, [32] although, as in humans, no single hypothesis has yet been proven to explain these observations.
It was interesting to note that there was an association between increasing faecal sieve score and C. fetus faecal-pat prevalence in cattle. This variable refers to the presence of undigested grains or long fibres in the diet, the presence of which suggests impaired ruminal digestion and the existence of a subacute ruminal acidosis [19]. This observation would support the hypothesis that diet is in part a determinant of C. fetus faecal-pat prevalence.
In the case of cattle C. fetus faecal-pat prevalence, there appeared to be significant variation at a relatively large spatial scale (zone), even after adjusting for small-scale spatial variation (farm) with a higher prevalence observed in farms situated in zone 3 (South Eastern Lancashire: OR 6·64, 95% CI 1·67–26·5, P = 0·01) and zone 2 (North East Lancashire: OR 2·0, 95% CI 0·53–7·6, P = 0·31) compared to farms in zone 1 (Fylde) although this was not statistically significant in the case of zone 2. This may be a reflection of the increased biodiversity likely to exist in the foothills and valleys of the Pennine uplands (zones 2 and 3) compared to the Fylde (zone 1) a low-lying predominately grassland and arable area. In the case of C. jejuni, we found greater genetic diversity in isolates from the Pennine areas compared to the Fylde and hypothesized that this may be a reflection of biodiversity with acquisition of C. jejuni from wildlife occurring to a larger extent in these areas compared to the Fylde. We suggest a similar state may pertain to C. fetus [17, 33].
This study demonstrates that C. fetus is commonly found in cattle and sheep faeces, and that seasonal and management factors associated with its detection in faeces could be identified. Given C. fetus is an important human [5] and animal pathogen [34] further work to investigate the molecular epidemiology of C. fetus in cattle and sheep is warranted in order to inform future interventions.
ACKNOWLEDGEMENTS
The work in this study was funded by the UK Department of Environment Food and Rural Affairs Veterinary Training and Research Initiative.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Silva J, et al. Campylobacter spp. as a foodborne pathogen: a review. Frontiers in Microbiology 2011; 2: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adak GK, et al. Disease risks from foods, England and Wales, 1996–2000. Emerging Infectious Diseases 2005; 11: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lastovica AJ, Allos BM. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and Campylobacter coli. In: Nachamkin I, Szymanski CM, Blaser MJ, eds. Campylobacter, 3rd edn. Washington DC: ASM Press, 2008, pp. 123–150. [Google Scholar]
- 4.Wagenaar JA, et al. Comparative study using amplified fragment length polymorphism fingerprinting, PCR genotyping, and phenotyping to differentiate Campylobacter fetus strains isolated from animals. Journal of Clinical Microbiology 2001; 39: 2283–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holst E, et al. Bacterial vaginosis: microbiological and clinical findings. European Journal of Clinical Microbiology 1987; 6: 536–541. [DOI] [PubMed] [Google Scholar]
- 6.Miki K, et al. Infective tricuspid valve endocarditis with pulmonary emboli caused by Campylobacter fetus after tooth extraction. Internal Medicine 2005; 44: 1055–1059. [DOI] [PubMed] [Google Scholar]
- 7.Rao GG, et al. Campylobacter fetus infections in two patients with AIDS. Journal of Infection 1990; 20: 170–172. [DOI] [PubMed] [Google Scholar]
- 8.Klein BS, et al. Campylobacter infection associated with raw milk. An outbreak of gastroenteritis due to Campylobacter jejuni and thermotolerant Campylobacter fetus subsp fetus. Journal of American Medical Association 1986; 255: 361–364. [DOI] [PubMed] [Google Scholar]
- 9.Rennie RP, et al. Campylobacter fetus diarrhea in a Hutterite colony: epidemiological observations and typing of the causative organism. Journal of Clinical Microbiology 1994; 32: 721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tremblay C, Gaudreau C, Lorange M. Epidemiology and antimicrobial susceptibilities of 111 Campylobacter fetus subsp. fetus strains isolated in Quebec, Canada, from 1983 to 2000. Journal of Clinical Microbiology 2003; 41: 463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser MJ, et al. Survival of Campylobacter fetus subsp. jejuni in biological milieus. Journal of Clinical Microbiology 1980; 11: 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFadyean J, Stockman S. Epizootic abortion. Appendix to Part III: Abortion in sheep. In: Report of the Departmental Committee Appointed by the Ministry of Agriculture and Fisheries, UK, 1913. London: His Majesty's Stationary Office, pp. 1–23. [Google Scholar]
- 13.UK Department of Food and Rural Affairs. Veterinary Investigation Surveillance Report. Yearly Trends 2004–2011: Sheep (http://vla.defra.gov.uk/reports/docs/rep_vida_sheep04_11.pdf). Accessed 12 November 2012.
- 14.Baird G, Gonzalez L. Campylobacter and Salmonella Abortion in Sheep. Moredun Foundation Newsheets, 2009, pp. 1–8. Galashiels: Midlothian Miegle Ltd. [Google Scholar]
- 15.Penner JL. The genus Campylobacter: a decade of progress. Clinical Microbiology Review 1988; 1: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grove-White DH, et al. Temporal and farm-management-associated variation in the faecal-pat prevalence of Campylobacter jejuni in ruminants. Epidemiology and Infection 2010; 138: 549–558. [DOI] [PubMed] [Google Scholar]
- 17.Grove-White DH, et al. Molecular epidemiology and genetic diversity of Campylobacter jejuni in ruminants. Epidemiology and Infection 2011; 139: 1661–1671. [DOI] [PubMed] [Google Scholar]
- 18.Hughes J. A system for assessing cow cleanliness. In Practice 2001; 23: 517–524. [Google Scholar]
- 19.Grove-White DH. Rumen health care in the dairy cow. In Practice 2004; 26: 88–95. [Google Scholar]
- 20.González I, et al. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Letters in Applied Microbiology 2000; 30: 207–212. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. Journal of Clinical Microbiology 2002; 40: 4744–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez I, et al. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. Journal of Clinical Microbiology 1997; 35: 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linton D, Owen RJ, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Research in Microbiology 1996; 147: 707–718. [DOI] [PubMed] [Google Scholar]
- 24.Van Bergen MA, et al. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. Journal of Clinical Microbiology 2005; 43: 5888–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan MS, Ventress N. Sequence typing analysis and retrieval system (http://neelix.molbiol.ox.ac.uk:8080/userweb/mchan/stars/index.html). University of Oxford, 2001. [Google Scholar]
- 26.Blaser MJ. Campylobacter fetus – emerging infection and model system for bacterial pathogenesis at mucosal surfaces. Clinical Infectious Disease 1998; 27: 256–258. [DOI] [PubMed] [Google Scholar]
- 27.Lawson A MLST analysis of Campylobacter fetus subsp. fetus isolates from humans suggests that sequence types have distinct aetiologies. Proceedings of CRL Campylobacter Workshop. Uppsala, Sweden: National Veterinary Institute, 2009. [Google Scholar]
- 28.Nylen G, et al. The seasonal distribution of Campylobacter infection in nine European countries and New Zealand. Epidemiology and Infection 2002; 128: 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen F, et al. Influence of season and geography on Campylobacter jejuni and C. coli subtypes in housed broiler flocks reared in Great Britain. Applied Environmental Microbiology 2011; 77: 3741–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley KN, et al. Seasonal variation of thermophilic campylobacters in lambs at slaughter. Journal of Applied Microbiology 1998; 84: 1111–1116. [DOI] [PubMed] [Google Scholar]
- 31.Kovats RS, et al. Climate variability and campylobacter infection: an international study. International Journal of Biometeorology 2005; 49: 207–214. [DOI] [PubMed] [Google Scholar]
- 32.Stanley KN, Jones K. Cattle and sheep farms as reservoirs of Campylobacter. Journal of Applied Microbiology 2003; 94 (Suppl.): 104S–113S. [DOI] [PubMed] [Google Scholar]
- 33.Luechtefeld NA, et al. Isolation of Campylobacter fetus subsp. jejuni from migratory waterfowl. Journal of Clinical Microbiology 1980; 12: 406–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Animal Health and Veterinary Laboraories Agency. The Non Statutory Zoonosis Report. Annual Report, 2011. (http://www.defra.gov.uk/ahvla-en/files/pub-zoo-ann11.pdf). Accessed 28 May 2013.