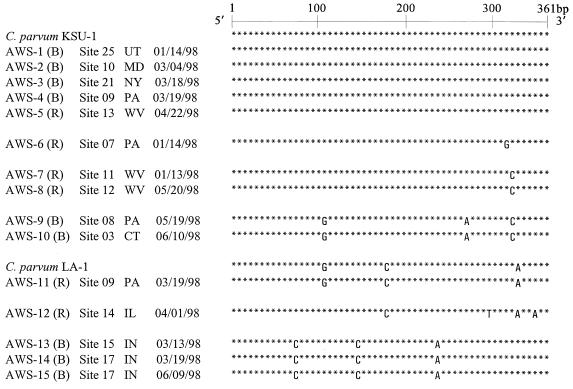
FIG. 1.
DNA sequence alignment of hsp70 CC-PCR products of infectious C. parvum recovered from raw (R) and filter backwash (B) water samples. States (postal abbreviations) from and dates (month/day/year) on which samples were recovered are indicated. C. parvum hsp70 PCR products were 361 bp long (18), and the C. parvum KSU-1 hsp70 sequence was the reference sequence (nucleotides [nt] 2423 to 2783). C. parvum LA-1 served as the laboratory quality control strain. Nucleotide substitutions from the 5′ end were as follows: nt 2507, T→C; nt 2528, A→G; nt 2563, T→C; nt 2591, T→C; nt 2646, C→A; nt 2682, G→A; nt 2714, C→T; nt 2731, A→G; nt 2740, T→C; nt 2747, G→A; and nt 2762, G→A. GenBank accession numbers for C. parvum sequences obtained in this study are provided in Materials and Methods.