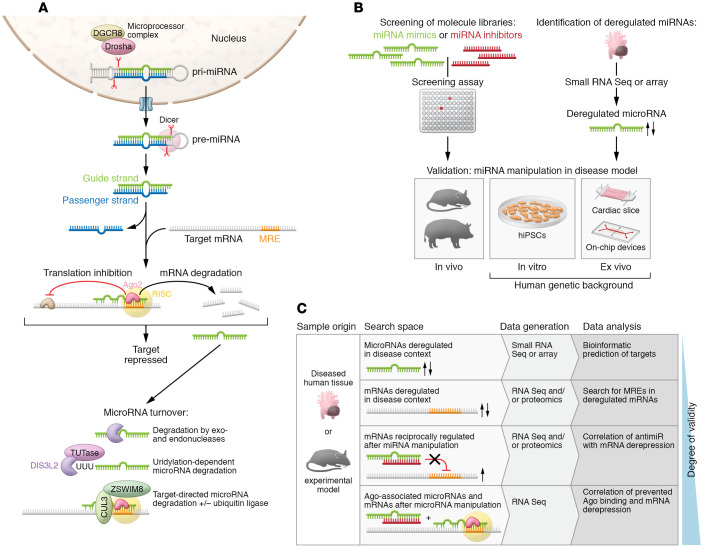
Figure 1. MicroRNA life cycle, and identification of microRNAs and their targets.
(A) Canonical processing, functional activation, mechanism of action, and degradation pathways of microRNAs. Canonical microRNA biogenesis starts from larger hairpin RNA molecules (pri-miRNAs), which are generated by RNA Pol II transcription of microRNA genes or clusters, or which occur as part of introns. A microprocessor complex that contains the endonuclease Drosha, the DiGeorge critical region 8 protein (DGCR8), and other factors then cleaves these pri-miRNAs. The resulting pre-miRNA is exported to the cytoplasm, where the nuclease Dicer tailors it to 21 to 22 nucleotides in length. There are also noncanonical mechanisms of microRNA biogenesis, some of which bypass the microprocessor complex or Dicer. After processing to a duplex of 21–22 nucleotides in length each, one strand, termed the guide strand, becomes part of the RNA-induced silencing complex (RISC), whereas the passenger strand (or *-strand) undergoes accelerated degradation. If both strands are maintained, they can adopt individual functions, as demonstrated for cardiovascular miR-21 and miR-126 (11, 12). Another exception are microRNA strands that localize to the nucleus, where they function in unusual manners (12, 13). Degradation of microRNAs involves exonucleases XRN-1, PNPase old-35, and RRP41 (17) or the endonuclease Tudor-SN (154). The nuclease DIS3L2 degrades a subset of microRNAs after modification by terminal uridyltransferases (TUTases) (155). Mechanisms of target-directed microRNA degradation (TDMD) have been resolved, including the involvement of ubiquitin ligases (25, 26). (B) Routes toward the identification and validation of disease-relevant cardiovascular microRNAs. (C) Approaches for the identification of microRNA targets.