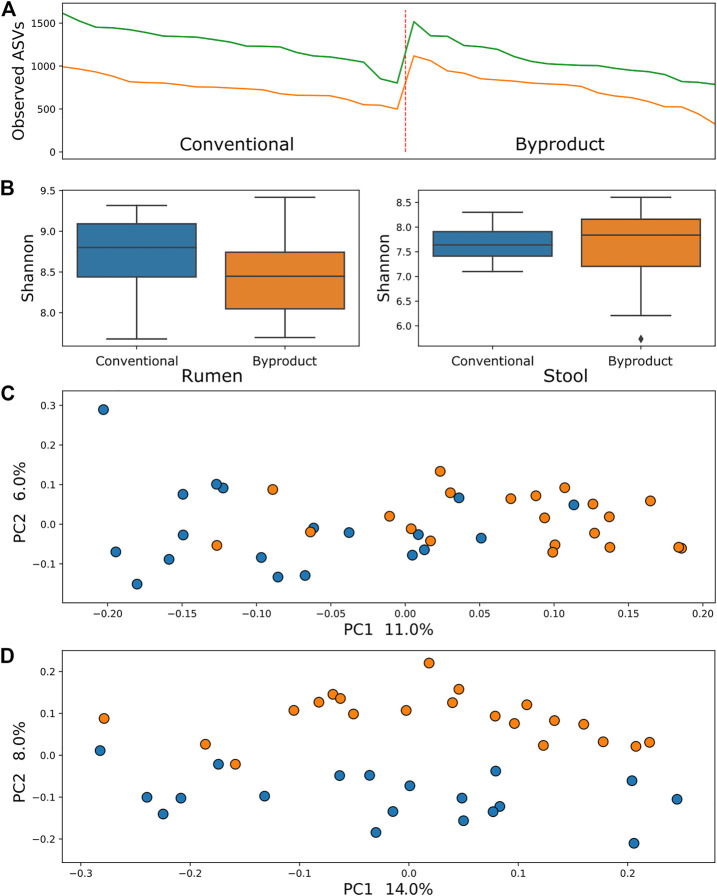
FIGURE 1.
(A) Comparison between observed ASV (amplicon sequence variants) metric between treatment groups showed no significant difference in the bacterial population of animals submitted to different diets in both rumen (green line) and stool (yellow line). (B) Shannon index comparisons showing a significant difference (p < 0.01) in the richness of bacteria from the rumen microbiome. (C) PCoA using the rumen microbiome unweighted UniFrac distance showing a tendency of clustering of samples from conventional group (blue) and B (orange). (D) PCoA using the stool microbiome unweighted UniFrac distance showing an almost linear separation of samples from animals fed conventional diet (blue) and by-product diet (orange).