Figure 1. The TPR domain of TSK specifically interacts with the N-terminal tail of the H3.1 variant.
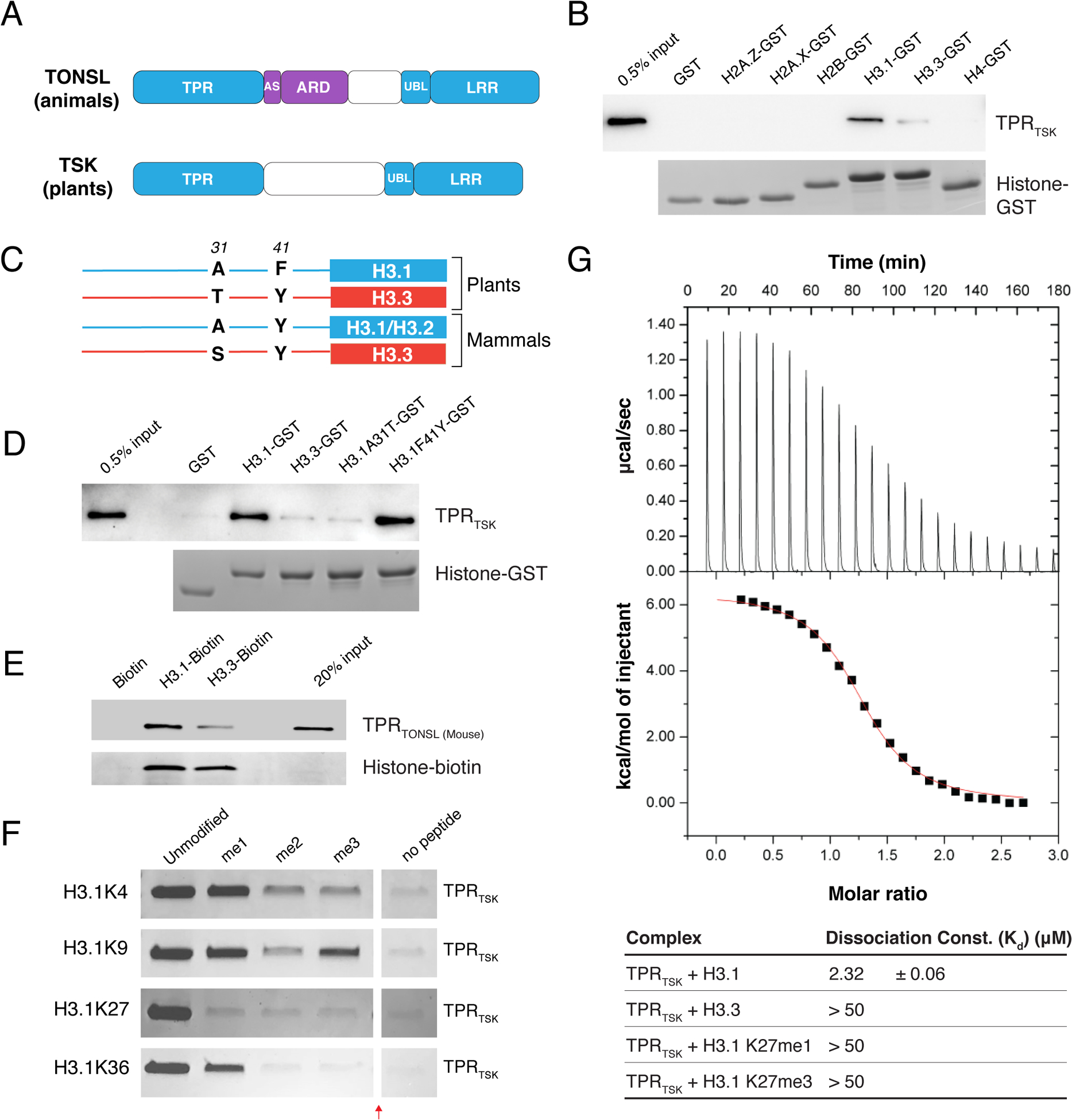
(A) Domain architecture of animal and plant TONSL/TSK. TPR: Tetratricopeptide Repeats, AS: Acidic Sequence, ARD: Ankyrin Repeat Domain, UBL: Ubiquitin-like, LRR: Leucine-Rich Repeats. Conserved domains are shown in blue. (B) Pull-down assay using TPRTSK and GST tagged with the N-terminal tails of histones H2A.Z, H2A.X, H2B, H3.1, H3.3 and H4 from plants. (C) Representation of plant and mammalian H3.1/H3.2 (blue) and H3.3 (red) H3 variants. Thin lines and blocks represent the histone tails and cores, respectively, and numbers indicate amino acid positions in H3. (D) Peptide pull-down assay using plant TPRTSK and GST tagged with the tails of histones H3.1, H3.3, H3.1A31T and H3.1F41Y. (E) Peptide pull-down assay using mouse TPRTONSL and biotin-tagged histones H3.1 and H3.3 (full-length proteins) from mammals. (F) Peptide pull-down assay using plant TPRTSK and methylated peptides at K4, K9, K27 and K36 of H3.1 (a.a. 1–45). The red arrow indicates a gel lane that was removed. (G) ITC assay using plant TPRTSK and different H3 peptides.
