Figure 4. H3.1 is required to mediate genomic instability in atxr5/6 mutants.
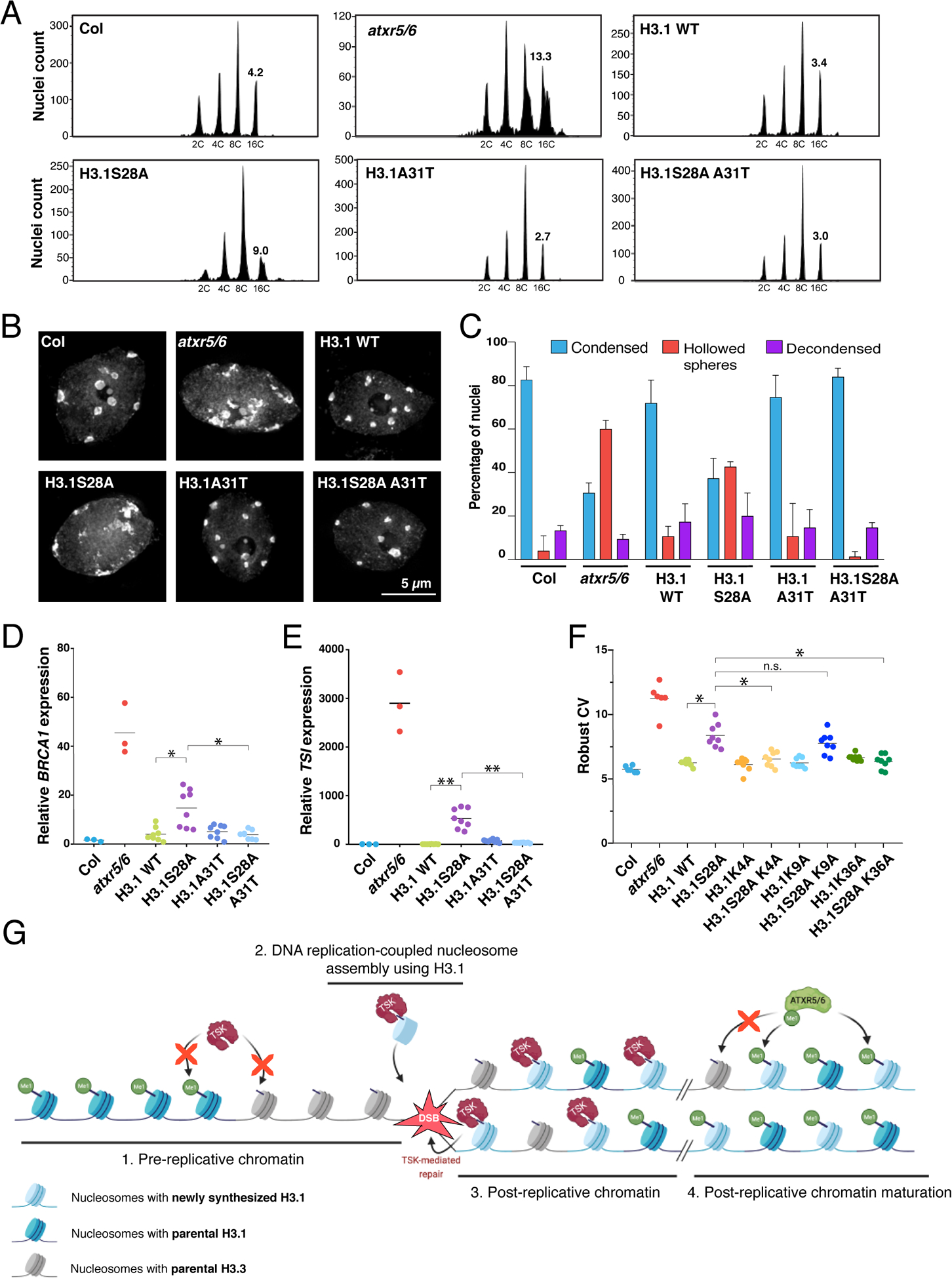
(A) Flow cytometry of leaf nuclei. Numbers below the peaks indicate ploidy, and those above indicate rCV. (B) Leaf nuclei of Col, atxr5/6, and first-generation (T1) H3.1 lines stained with DAPI. (C) Quantification from nuclei in B. Error bars indicate SEM. (D, E) RT-qPCR of BRCA1 and TSI. Horizontal bars indicate the mean. Welch’s ANOVA followed by the Dunnett’s T3 test: * p < 0.05, ** p < 0.001. (F) rCV for 16C nuclei obtained by flow cytometry. For Col and atxr5/6, each dot represents a biological replicate. For the H3.1 lines, each dot represents one T1 plant. Horizontal bars indicate the mean. Welch’s ANOVA followed by the Dunnett’s T3 test: * p < 0.05, n.s. = not significantly different. (G) Model depicting the interplay between H3.1, TSK and ATXR5/6 during replication. Step 1. TSK cannot interact with chromatin containing H3.3K27me0 or H3.1K27me1. Step 2. Newly synthesized H3.1 (H3.1K27me0) in complex with TSK are inserted at replication forks. Step 3. DSBs caused by broken replication forks are repaired by TSK. Step 4. Mono-methylation of newly inserted H3.1 (but not H3.3) at K27 by ATXR5/6 prevents binding of TSK.
